Team:Newcastle/2 September 2010
From 2010.igem.org
(Difference between revisions)
Swoodhouse (Talk | contribs) (→Materials and Protocol) |
Swoodhouse (Talk | contribs) (→Transformation of Bacillius subtilis 168 with pGFP-rrnB containing filamentous cell part) |
||
| Line 5: | Line 5: | ||
N.B. During autoclaving we loose some volume of the solution. | N.B. During autoclaving we loose some volume of the solution. | ||
| - | = | + | =''yneA''= |
| - | == | + | ==Aims== |
| - | + | ||
| + | The aim of the experiment is to test if the filamentous cell part was cloned into pGFP-rrnB successfully. | ||
| - | [[Image: | + | ==Materials and protocols== |
| + | |||
| + | #Please refer to: [[Team:Newcastle/Qiagen_Minipreps#Plasmid_extraction| Plasmid extraction]]. | ||
| + | #Please refer to:[[Team:Newcastle/Restriction_digests|Restriction digest]]. We used Nhe1 and Spe1 to remove the ''yneA'' from pGFP-rrnB. | ||
| + | #Please refer to:[[Team:Newcastle/Gel_electrophoresis| Gel electrophoresis]] for running all the digested plasmid fragments. | ||
| + | |||
| + | ==Results== | ||
| + | |||
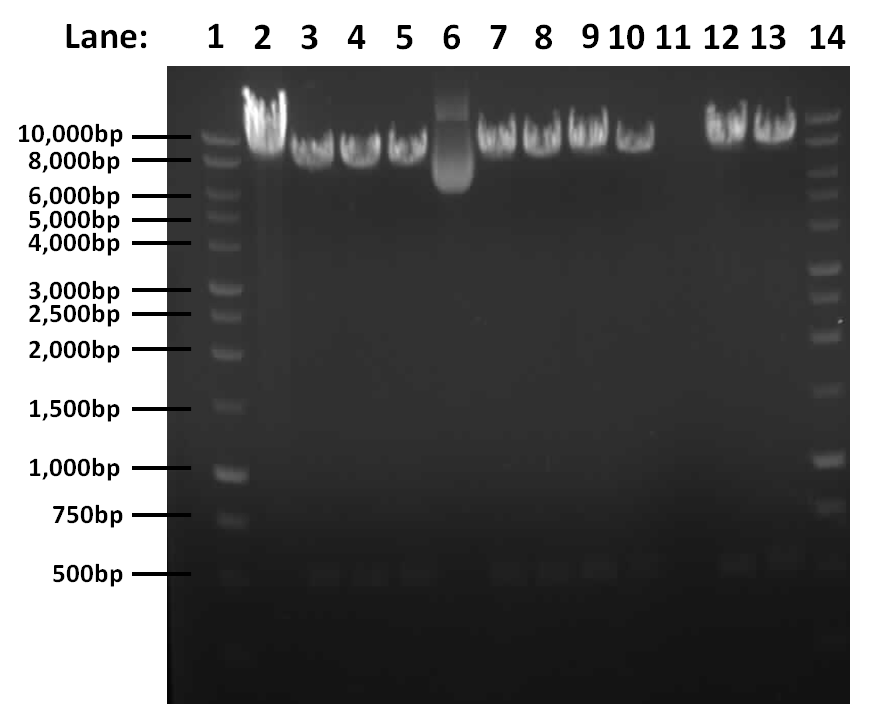
| + | [[Image:03.09.10.png#file|400px]] | ||
| + | |||
| + | '''Figure 1''': Gel electrophoresis result for restriction digest of pGFPrrnB and ''yneA'' with Nhe1 and Spe1. | ||
| + | |||
| + | * '''Lane 1''': 1kb DNA ladder | ||
| + | * '''Lane 2''': Tube 1 | ||
| + | * '''Lane 3''': Tube 2 | ||
| + | * '''Lane 4''': Tube 3 | ||
| + | * '''Lane 5''': Tube 4 | ||
| + | * '''Lane 6''': Tube 5 | ||
| + | * '''Lane 7''': Tube 6 | ||
| + | * '''Lane 8''': Tube 7 | ||
| + | * '''Lane 9''': Tube 8 | ||
| + | * '''Lane 10''': Tube 9 | ||
| + | * '''Lane 11''': Tube 10 | ||
| + | * '''Lane 12''': Tube 11 | ||
| + | * '''Lane 13''': Tube 12 | ||
| + | * '''Lane 14''': 1kb DNA ladder | ||
| + | |||
| + | ==Conclusion== | ||
| + | |||
| + | The results show that the digest works, there is a band at approximately 541bp corresponding to ''yneA'' and a band at approximately 8.4kbp corresponding to pGFPrrnb in lanes 2, 3, 4, 6, 7, 8, 9, 11 and 12. | ||
| - | |||
| - | |||
| - | |||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Revision as of 14:20, 27 October 2010

| |||||||||||||
| |||||||||||||
Contents |
pH changing for HEPES
We prepared 100mM of HEPES (it is a dibasic compound) at pH 7.0 and sent it for autoclaving. N.B. During autoclaving we loose some volume of the solution.
yneA
Aims
The aim of the experiment is to test if the filamentous cell part was cloned into pGFP-rrnB successfully.
Materials and protocols
- Please refer to: Plasmid extraction.
- Please refer to:Restriction digest. We used Nhe1 and Spe1 to remove the yneA from pGFP-rrnB.
- Please refer to: Gel electrophoresis for running all the digested plasmid fragments.
Results
Figure 1: Gel electrophoresis result for restriction digest of pGFPrrnB and yneA with Nhe1 and Spe1.
- Lane 1: 1kb DNA ladder
- Lane 2: Tube 1
- Lane 3: Tube 2
- Lane 4: Tube 3
- Lane 5: Tube 4
- Lane 6: Tube 5
- Lane 7: Tube 6
- Lane 8: Tube 7
- Lane 9: Tube 8
- Lane 10: Tube 9
- Lane 11: Tube 10
- Lane 12: Tube 11
- Lane 13: Tube 12
- Lane 14: 1kb DNA ladder
Conclusion
The results show that the digest works, there is a band at approximately 541bp corresponding to yneA and a band at approximately 8.4kbp corresponding to pGFPrrnb in lanes 2, 3, 4, 6, 7, 8, 9, 11 and 12.
 
|
 "
"