Team:Newcastle/2 July 2010
From 2010.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | |||
| - | === | + | '''2 July 2010''' |
| + | |||
| + | =Urease Test= | ||
| + | |||
| + | ==Aims== | ||
The aim of this experiment was to determine if ''Bacillus subtilis'' 168 is able to produce urease and degrade urea. | The aim of this experiment was to determine if ''Bacillus subtilis'' 168 is able to produce urease and degrade urea. | ||
| - | + | ==Procedures== | |
The experiment was performed on 01.07.10. Please refer to [[Team:Newcastle/Urease test| Urease test]]. | The experiment was performed on 01.07.10. Please refer to [[Team:Newcastle/Urease test| Urease test]]. | ||
| - | + | ==Results== | |
|[[Image:Newcastle_urease_test_020710.png|400px ]] | |[[Image:Newcastle_urease_test_020710.png|400px ]] | ||
| Line 16: | Line 19: | ||
# Test 2 (Duplicate) - Pink-red color | # Test 2 (Duplicate) - Pink-red color | ||
| - | + | ==Discussion== | |
''B. subtilis'' 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color. | ''B. subtilis'' 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color. | ||
| - | + | ==Conclusion== | |
The negative control did not turn pink-red color, therefore indicating that no contamination had occurred. | The negative control did not turn pink-red color, therefore indicating that no contamination had occurred. | ||
| - | + | =LacI BioBrick Construction= | |
| - | + | ==Aims== | |
*To use PCR to extract ''lacI'' (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP). | *To use PCR to extract ''lacI'' (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP). | ||
| - | + | ==Materials== | |
None | None | ||
| - | + | ==Protocol== | |
* Transformed ''E. coli'' DH5alpha are stored in a refrigerator over the weekend. | * Transformed ''E. coli'' DH5alpha are stored in a refrigerator over the weekend. | ||
| - | + | ==Inference== | |
*''Ecoli'' DH5alpha are kept viable by the colder temperature ready for plasmid extraction after the weekend. | *''Ecoli'' DH5alpha are kept viable by the colder temperature ready for plasmid extraction after the weekend. | ||
| - | + | =Competent ''E. coli'' Production= | |
| - | + | ==Aims== | |
*To make a stock of competent ''E. coli'' (DH5alpha strain) ready for transformation. | *To make a stock of competent ''E. coli'' (DH5alpha strain) ready for transformation. | ||
| - | + | ==Materials== | |
*Liquid culture of ''E. coli'' (DH5alpha strain). | *Liquid culture of ''E. coli'' (DH5alpha strain). | ||
| - | + | ==Protocol== | |
*Cause ''E. coli'' (DH5alpha strain) to become [[Team:Newcastle/E. coli Competence|competent]]. | *Cause ''E. coli'' (DH5alpha strain) to become [[Team:Newcastle/E. coli Competence|competent]]. | ||
| - | + | ==Inference== | |
*We created a stock of competent ''E. coli'' (DH5alpha strain) that an be used for future transformations to grow up/replicate plasmids. | *We created a stock of competent ''E. coli'' (DH5alpha strain) that an be used for future transformations to grow up/replicate plasmids. | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Revision as of 10:48, 11 August 2010

| |||||||||||||
| |||||||||||||
2 July 2010
Contents |
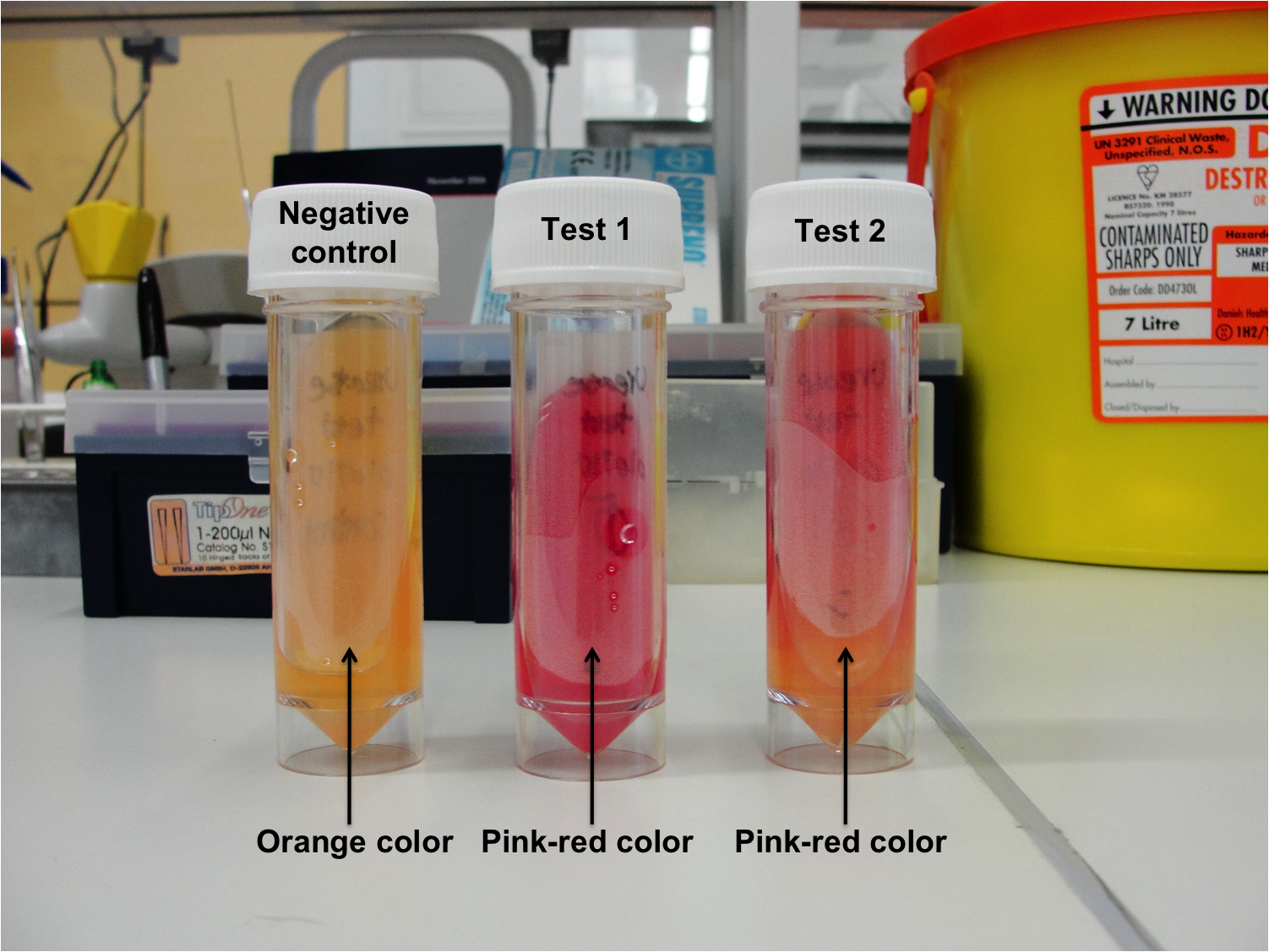
Urease Test
Aims
The aim of this experiment was to determine if Bacillus subtilis 168 is able to produce urease and degrade urea.
Procedures
The experiment was performed on 01.07.10. Please refer to Urease test.
Results
Figure 1: Urease test results
- Negative control - No color change (Orange color)
- Test 1 (Duplicate) - Pink-red color
- Test 2 (Duplicate) - Pink-red color
Discussion
B. subtilis 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color.
Conclusion
The negative control did not turn pink-red color, therefore indicating that no contamination had occurred.
LacI BioBrick Construction
Aims
- To use PCR to extract lacI (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP).
Materials
None
Protocol
- Transformed E. coli DH5alpha are stored in a refrigerator over the weekend.
Inference
- Ecoli DH5alpha are kept viable by the colder temperature ready for plasmid extraction after the weekend.
Competent E. coli Production
Aims
- To make a stock of competent E. coli (DH5alpha strain) ready for transformation.
Materials
- Liquid culture of E. coli (DH5alpha strain).
Protocol
- Cause E. coli (DH5alpha strain) to become competent.
Inference
- We created a stock of competent E. coli (DH5alpha strain) that an be used for future transformations to grow up/replicate plasmids.
 
|
 "
"