Team:Newcastle/2 July 2010
From 2010.igem.org
(→Urease test) |
Shethharsh08 (Talk | contribs) |
||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | |||
| - | === | + | '''2 July 2010''' |
| + | |||
| + | =Urease Test= | ||
| + | |||
| + | ==Aims== | ||
The aim of this experiment was to determine if ''Bacillus subtilis'' 168 is able to produce urease and degrade urea. | The aim of this experiment was to determine if ''Bacillus subtilis'' 168 is able to produce urease and degrade urea. | ||
| - | == | + | ==Procedures== |
| - | The experiment was performed on 01.07.10. | + | The experiment was performed on 01.07.10. Please refer to [[Team:Newcastle/Urease test| Urease test]]. |
| - | + | ==Results== | |
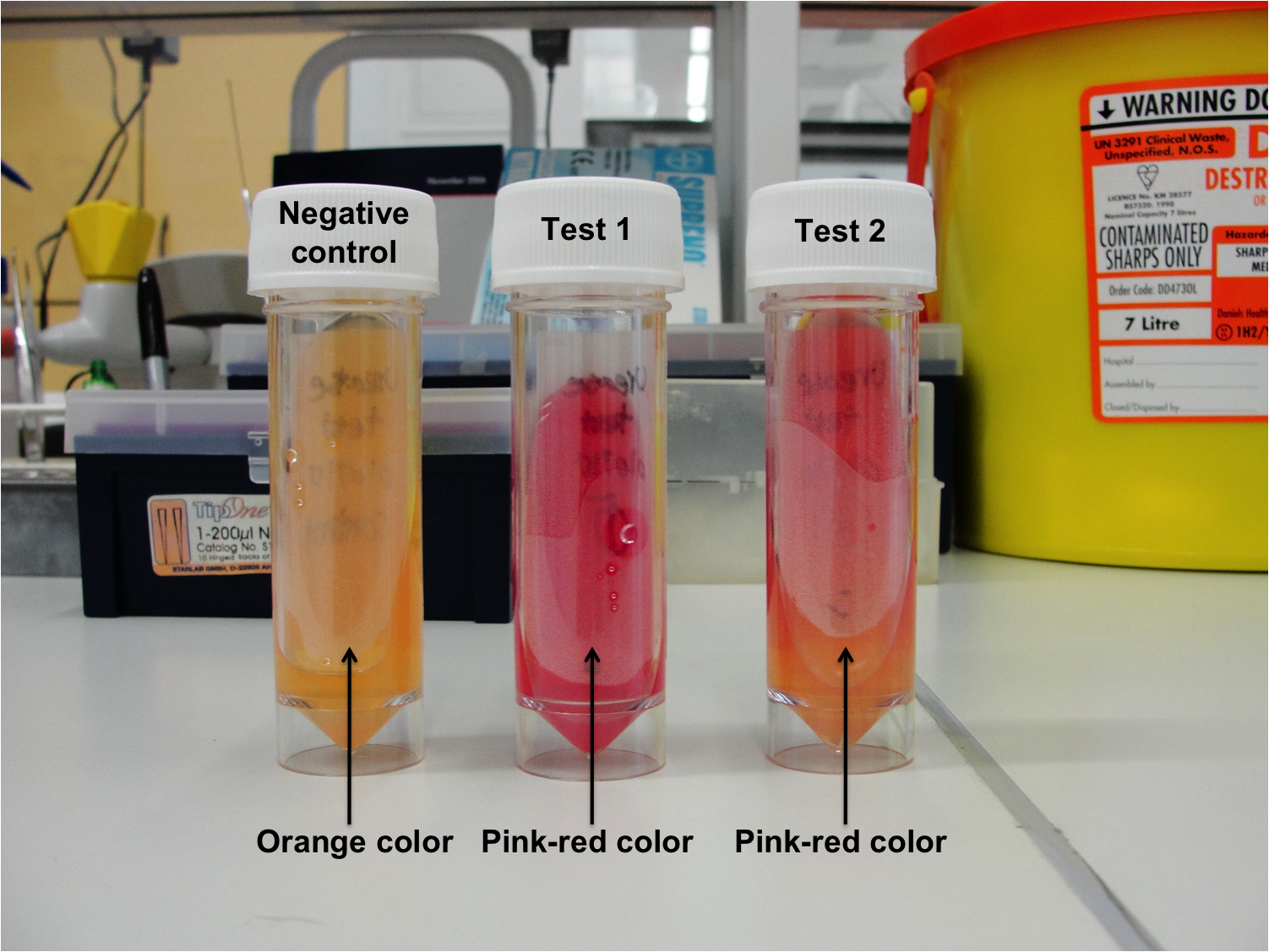
|[[Image:Newcastle_urease_test_020710.png|400px ]] | |[[Image:Newcastle_urease_test_020710.png|400px ]] | ||
| Line 16: | Line 19: | ||
# Test 2 (Duplicate) - Pink-red color | # Test 2 (Duplicate) - Pink-red color | ||
| - | + | ==Discussion== | |
| - | ''B. subtilis'' 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color. | + | ''B. subtilis'' 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color. The negative control did not turn pink-red color, therefore indicating that no contamination had occurred. |
| - | + | ||
| - | + | ||
| - | The negative control did not turn pink-red color, therefore indicating that no contamination had occurred. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==Conclusion== |
| - | + | From the experiment it can be concluded that ''Bacillus subtilis'' 168 produces urease enzyme in order to break down urea and produces ammonium and carbonate ions. | |
| - | |||
| - | |||
| + | =Competent ''E. coli'' DH5α Production= | ||
| + | ==Aims== | ||
| + | *To make a stock of competent ''E. coli'' (DH5α strain) ready for transformation. | ||
| - | + | ==Materials and Protocol== | |
| + | Please refer to:[[Team:Newcastle/E. coli Competence|competent]] for materials and protocol. | ||
| + | ==Inference== | ||
| + | *We created a stock of competent ''E. coli'' (DH5α strain) that an be used for future transformations to grow up/replicate plasmids. | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 14:19, 25 October 2010

| |||||||||||||
| |||||||||||||
2 July 2010
Contents |
Urease Test
Aims
The aim of this experiment was to determine if Bacillus subtilis 168 is able to produce urease and degrade urea.
Procedures
The experiment was performed on 01.07.10. Please refer to Urease test.
Results
Figure 1: Urease test results
- Negative control - No color change (Orange color)
- Test 1 (Duplicate) - Pink-red color
- Test 2 (Duplicate) - Pink-red color
Discussion
B. subtilis 168 is able to produce urease, therefore, urea which is the substrate and can be found in the agar was degraded, which results in ammonia building. The ammonia makes the media alkaline and therefore the indicator phenol red will change from orange color to pink-red color. The negative control did not turn pink-red color, therefore indicating that no contamination had occurred.
Conclusion
From the experiment it can be concluded that Bacillus subtilis 168 produces urease enzyme in order to break down urea and produces ammonium and carbonate ions.
Competent E. coli DH5α Production
Aims
- To make a stock of competent E. coli (DH5α strain) ready for transformation.
Materials and Protocol
Please refer to:competent for materials and protocol.
Inference
- We created a stock of competent E. coli (DH5α strain) that an be used for future transformations to grow up/replicate plasmids.
 
|
 "
"