Team:Newcastle/28 July 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) (New page: {{Team:Newcastle/mainbanner}} ===Aims=== The aim of this experiment is to prove that ''Bacillus subtilis'' 168 can take up arginine from the media into the cell. ===Materials=== * pH str...) |
(→Plasmid Miniprep Experiment) |
||
| (65 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | ===Aims | + | [[Image:Newcastle_Lab_4.jpeg|260px|right]] |
| - | The aim of this experiment is to prove that ''Bacillus subtilis'' 168 | + | |
| + | =PCR Experiment= | ||
| + | |||
| + | ==Aims== | ||
| + | The aim of this experiment is to prove that ''Bacillus subtilis'' 168 and 3610 chromosomal DNA extraction worked by amplifying P''araE'' using PCR. | ||
| + | |||
| + | [[Image:Newcastle_Thermo.JPG|200px|thumb|right|Thermocycler]] | ||
| + | [[Image:Newcastle Loading Gel.jpg|200px|thumb|right|Loading the gel]] | ||
| + | [[Image:Newcastle Prep Chr Gel.jpg|200px|thumb|right|Gel]] | ||
| + | |||
| + | ==Materials and Protocol== | ||
| + | |||
| + | Please refer to: [[Team:Newcastle/PCR#GoTaq_PCR|GoTaq PCR protocol]]. | ||
| + | |||
| + | ==Result== | ||
| + | |||
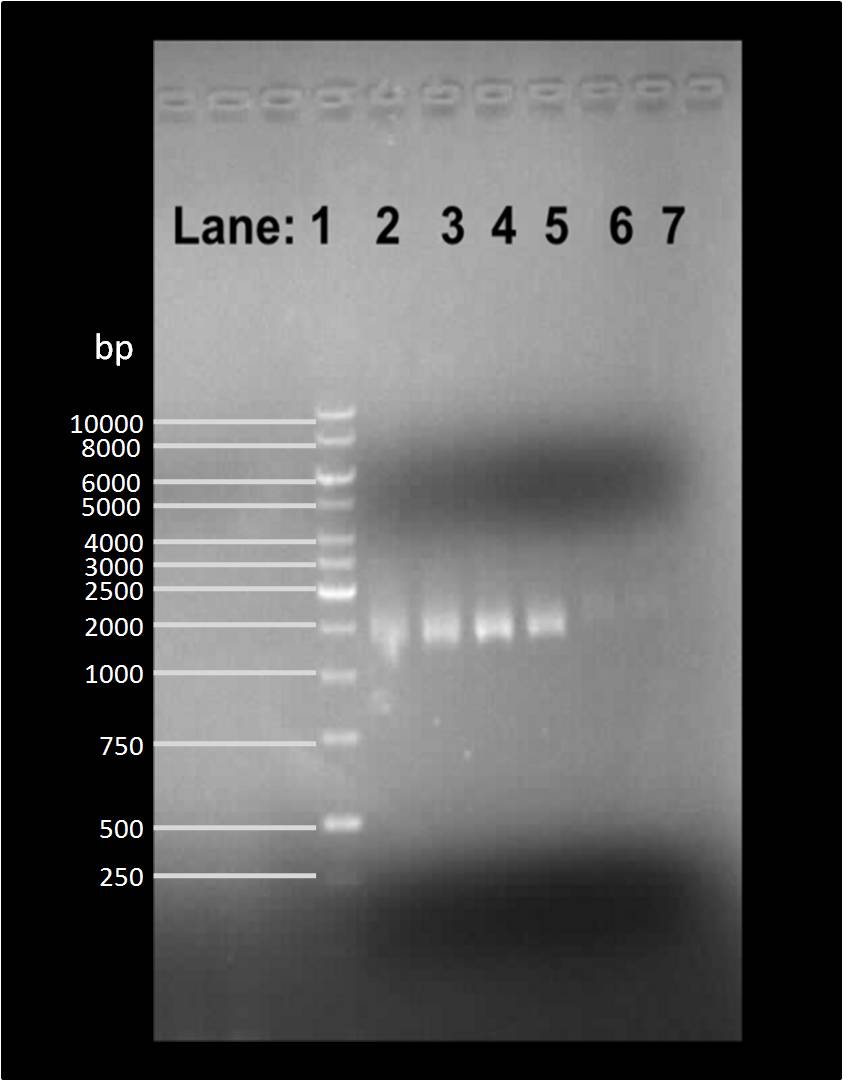
| + | [[Image:Gelpic2807.png|300px]] | ||
| + | |||
| + | '''Figure 1''': Gel electrophoresis of the PCR products | ||
| + | |||
| + | * '''Lane 1''': 1 Kb DNA ladder | ||
| + | * '''Lane 2''': ''B. subtilis'' 168 chromosomal DNA containing P''araE'' | ||
| + | * '''Lane 3''': ''B. subtilis'' 168 chromosomal DNA containing P''araE'' | ||
| + | * '''Lane 4''': ''B. subtilis'' 3610 chromosomal DNA containing P''araE'' | ||
| + | * '''Lane 5''': ''B. subtilis'' 3610 chromosomal DNA containing P''araE'' | ||
| + | |||
| + | ==Discussion== | ||
| + | We found bands in lanes 2, 3, 4 and 5 of around 200 bp in size which is an approximate size of the P''araE'' which is found on the chromosome of both ''B. subtilis'' 168 and 3610. | ||
| + | |||
| + | ==Conclusion== | ||
| + | This experiment proves that the DNA extraction from both ''B. subtilis'' 168 and 3610 done on 27th July, 2010 was successful. | ||
| + | |||
| + | =Plasmid Miniprep Experiment= | ||
| + | |||
| + | ==Aims== | ||
| + | The aim of this experiment is to extract plasmid DNA pSB1C3, [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] and plasmid containing ''lacI'' Biobrick from ''E. coli'' DH5α cells with the help of Qiagen miniprep kit and confirming the extraction with the help of nanodrop experiment. | ||
| + | |||
| + | ==Materials and Protocol== | ||
| + | [[Image:Newcastle_overnight_culture.jpg|200px|thumb|right]] | ||
| + | [[Image:Newcastle_plasmids.jpg|200px|thumb|right]] | ||
| + | [[Image:Newcastle_pSB1C3.jpg|200px|thumb|right]] | ||
| + | |||
| + | Please refer to: [[Team:Newcastle/Minipreps| Minipreps]] for Qiagen miniprep protocol and [[TeamNewcastleNanoDrop Spectrophotometer| Nanodrop Spectrophotometer]] for nanodrop protocol. | ||
| + | |||
| + | ==Result== | ||
| + | |||
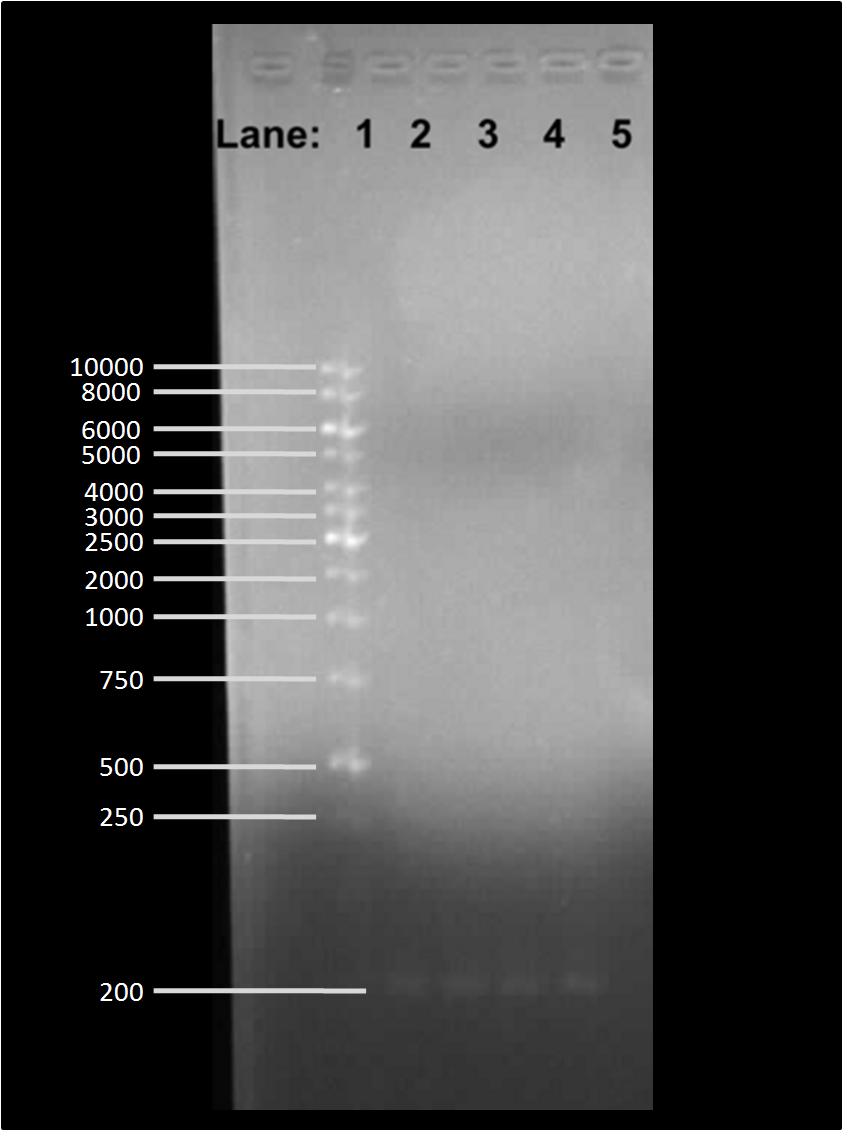
| + | [[Image:Gelpic28073.jpg|300px]] | ||
| + | |||
| + | '''Figure 2''': Gel electrophoresis of the plasmid after restriction digestion with EcoR1. | ||
| + | |||
| + | * '''Lane 1''': 1 kb DNA ladder | ||
| + | * '''Lane 2''': Extraction of pSB1C3 plasmid | ||
| + | * '''Lane 3''': Extraction of pSB1C3 plasmid | ||
| + | * '''Lane 4''': Extraction of plasmid containing ''lacI'' | ||
| + | * '''Lane 5''': Extraction of plasmid containing ''lacI'' | ||
| + | * '''Lane 6''': Extraction of [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] plasmid containing double terminator | ||
| + | * '''Lane 7''': Extraction of [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] plasmid containing double terminator | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{|border=1 | {|border=1 | ||
|- | |- | ||
| - | ! | + | !'''Lane 1''' |
| - | ! | + | !'''Lane 2''' |
| - | ! | + | !'''Lane 3''' |
| + | !'''Lane 4''' | ||
| + | !'''Lane 5''' | ||
| + | !'''Lane 6''' | ||
| + | !'''Lane 7''' | ||
|- | |- | ||
| - | | | + | |N/A |
| - | | | + | |29.9 µl/ml |
| - | | | + | |28.9 µl/ml |
| - | | | + | |34.0 µl/ml |
| - | + | |29.8 µl/ml | |
| - | + | |6.1 µl/ml | |
| - | + | |6.7 µl/ml | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
|} | |} | ||
| + | '''Table 1''': Nanodrop spectrophotometer experiment result. Table represents the amount of plasmid present in µl/ml quantity. | ||
| - | + | ==Discussion== | |
| - | + | We found bands in the lane 2, 3, 4, 5, and 6 showing the presence of plasmid in ''E. coli'' DH5α cells. | |
| - | + | However, the concentration was very low, therefore this will have to be repeated. | |
| - | + | ||
| - | + | ||
| + | ==Conclusion== | ||
| + | This experiment shows that there is plasmid present in the ''E. coli'' DH5α cells but they are present in a very low amount possibly due to the ommission of antibiotic in the media. | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 22:28, 27 October 2010

| |||||||||||||
| |||||||||||||
Contents |
PCR Experiment
Aims
The aim of this experiment is to prove that Bacillus subtilis 168 and 3610 chromosomal DNA extraction worked by amplifying ParaE using PCR.
Materials and Protocol
Please refer to: GoTaq PCR protocol.
Result
Figure 1: Gel electrophoresis of the PCR products
- Lane 1: 1 Kb DNA ladder
- Lane 2: B. subtilis 168 chromosomal DNA containing ParaE
- Lane 3: B. subtilis 168 chromosomal DNA containing ParaE
- Lane 4: B. subtilis 3610 chromosomal DNA containing ParaE
- Lane 5: B. subtilis 3610 chromosomal DNA containing ParaE
Discussion
We found bands in lanes 2, 3, 4 and 5 of around 200 bp in size which is an approximate size of the ParaE which is found on the chromosome of both B. subtilis 168 and 3610.
Conclusion
This experiment proves that the DNA extraction from both B. subtilis 168 and 3610 done on 27th July, 2010 was successful.
Plasmid Miniprep Experiment
Aims
The aim of this experiment is to extract plasmid DNA pSB1C3, [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] and plasmid containing lacI Biobrick from E. coli DH5α cells with the help of Qiagen miniprep kit and confirming the extraction with the help of nanodrop experiment.
Materials and Protocol
Please refer to: Minipreps for Qiagen miniprep protocol and Nanodrop Spectrophotometer for nanodrop protocol.
Result
Figure 2: Gel electrophoresis of the plasmid after restriction digestion with EcoR1.
- Lane 1: 1 kb DNA ladder
- Lane 2: Extraction of pSB1C3 plasmid
- Lane 3: Extraction of pSB1C3 plasmid
- Lane 4: Extraction of plasmid containing lacI
- Lane 5: Extraction of plasmid containing lacI
- Lane 6: Extraction of [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] plasmid containing double terminator
- Lane 7: Extraction of [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] with [http://partsregistry.org/Part:BBa_B0014 BBa_B0014] plasmid containing double terminator
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 |
|---|---|---|---|---|---|---|
| N/A | 29.9 µl/ml | 28.9 µl/ml | 34.0 µl/ml | 29.8 µl/ml | 6.1 µl/ml | 6.7 µl/ml |
Table 1: Nanodrop spectrophotometer experiment result. Table represents the amount of plasmid present in µl/ml quantity.
Discussion
We found bands in the lane 2, 3, 4, 5, and 6 showing the presence of plasmid in E. coli DH5α cells. However, the concentration was very low, therefore this will have to be repeated.
Conclusion
This experiment shows that there is plasmid present in the E. coli DH5α cells but they are present in a very low amount possibly due to the ommission of antibiotic in the media.
 
|
 "
"