Team:Newcastle/25 June 2010
From 2010.igem.org
(Difference between revisions)
RachelBoyd (Talk | contribs) (→Polymerase Chain Reaction protocol) |
Shethharsh08 (Talk | contribs) (→Result) |
||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | |||
| - | == | + | =Polymerase Chain Reaction= |
| - | + | ||
| - | == | + | ==Aim== |
| - | + | To amplify a desired DNA fragment by using a set of appropriate primers flanking the fragment. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==Materials and Protocol== |
| - | + | Please refer to: [[Team:Newcastle/PCR| PCR]] for materials required and protocol. | |
| - | + | ||
| - | + | ||
| - | + | ==Result== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
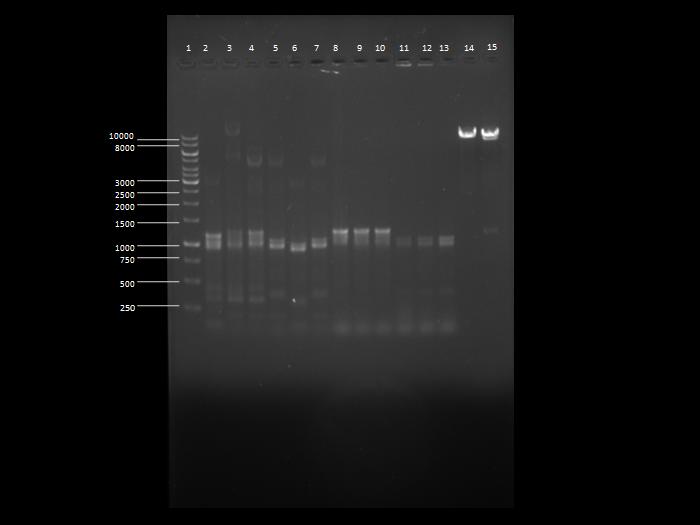
| - | + | [[Image:8062010.jpg|400px|center]] | |
| - | + | ||
| - | + | '''Image 1''': Image of the gel showing PCR products. | |
| - | + | ||
| + | * '''Lane 1''': 1kb DNA ladder | ||
| + | * '''Lane 2''': Sample 1 | ||
| + | * '''Lane 3''': Sample 2 | ||
| + | * '''Lane 4''': Sample 3 | ||
| + | * '''Lane 5''': Sample 4 | ||
| + | * '''Lane 6''': Sample 5 | ||
| + | * '''Lane 7''': Sample 6 | ||
| + | * '''Lane 8''': Sample 7 | ||
| + | * '''Lane 9''': Sample 8 | ||
| + | * '''Lane 10''': Sample 9 | ||
| + | * '''Lane 11''': Sample 10 | ||
| + | * '''Lane 12''': Sample 11 | ||
| + | * '''Lane 13''': Sample 12 | ||
| + | * '''Lane 14''': Sample 13 | ||
| + | * '''Lane 15''': Sample 14 | ||
| + | |||
| + | |||
| + | |||
| + | {{Team:Newcastle/footer}} | ||
Latest revision as of 13:54, 27 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Polymerase Chain Reaction
Aim
To amplify a desired DNA fragment by using a set of appropriate primers flanking the fragment.
Materials and Protocol
Please refer to: PCR for materials required and protocol.
Result
Image 1: Image of the gel showing PCR products.
- Lane 1: 1kb DNA ladder
- Lane 2: Sample 1
- Lane 3: Sample 2
- Lane 4: Sample 3
- Lane 5: Sample 4
- Lane 6: Sample 5
- Lane 7: Sample 6
- Lane 8: Sample 7
- Lane 9: Sample 8
- Lane 10: Sample 9
- Lane 11: Sample 10
- Lane 12: Sample 11
- Lane 13: Sample 12
- Lane 14: Sample 13
- Lane 15: Sample 14
 
|
 "
"