Team:Newcastle/25 August 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) (→PCR) |
Shethharsh08 (Talk | contribs) (→PCR) |
||
| Line 186: | Line 186: | ||
|Plasmid Vector | |Plasmid Vector | ||
|pSB1C3 | |pSB1C3 | ||
| - | | | + | |pSB1C3_for |
| - | | | + | |pSB1C3_rev |
|64 | |64 | ||
|2072 approx. | |2072 approx. | ||
| Line 195: | Line 195: | ||
|Plasmid Vector | |Plasmid Vector | ||
|pSB1C3 | |pSB1C3 | ||
| - | | | + | |pSB1C3_for |
| - | | | + | |pSB1C3_rev |
|65 | |65 | ||
|2072 approx. | |2072 approx. | ||
| Line 204: | Line 204: | ||
|Plasmid Vector | |Plasmid Vector | ||
|pSB1C3 | |pSB1C3 | ||
| - | | | + | |pSB1C3_for |
| - | | | + | |pSB1C3_rev |
|66 | |66 | ||
|2072 approx. | |2072 approx. | ||
Revision as of 14:00, 26 August 2010

| |||||||||||||
| |||||||||||||
Contents |
yneA
Gel Electrophoresis
Aim
To check if the digestion from yesterday worked.
Materials and Protocol
Please refer to gel electrophoresis for protocol.
Results
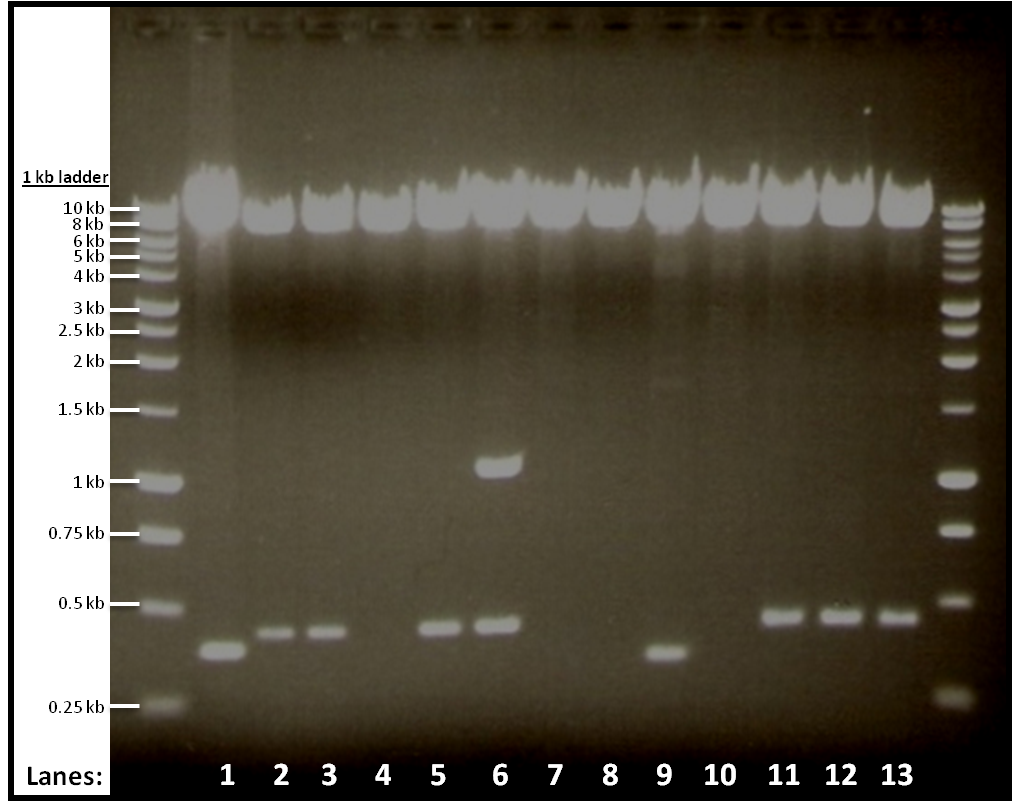
The result from gel electrophoresis:
Figure 1 shows the double digest of 12 tubes of pGFPrrnB and yneA.
- Lane 1: Vector only
- Lane 2: Tube 1
- Lane 3: Tube 2
- Lane 4: Tube 3
- Lane 5: Tube 4
- Lane 6: Tube 5
- Lane 7: Tube 6
- Lane 8: Tube 7
- Lane 9: Tube 8
- Lane 10: Tube 9
- Lane 11: Tube 10
- Lane 12: Tube 11
- Lane 13: Tube 12
Discussion
The bands we got from the gel shows that digestion in tubes 1, 2, 4, 10, 11 and 12 worked.
Conclusion
We use the digested products from the six tubes that worked for ligation.
Transformation of B. subtilis
Aim
To transform yneA into competent B. subtilis.
Materials and Protocol
Please refer to transformation of B. subtilis.
Results and Conclusion
Please refer to results in tomorrow's lab book.
Transformation
Aim
To transform pGFPrrnB and yneA into E. coli.
Materials and Protocol
Please refer to transformation of E. coli.
Results and Conclusion
Please refer to tomorrow's lab book.
PCR
Aim
To amplify the DNA pSB1C3 using RocF primer.
Materials and Protocol
Please refer to PCR.
Conclusion
Continue with PCR purification.
PCR Purification
Aim
To remove unwanted primers, taq polymerase, buffer and salts to obtain pure DNA.
Materials and Protocol
Please refer to PCR purification.
Digestion
Aim
To digest the PCR products of pSB1C3 and yneA from PCR purification.
Materials and Protocol
Please refer to restriction digest.
Gel extraction
Aim
To purify the DNA of yneA and pSB1C3 by extracting the bands from the gel after running gel electrophoresis. Concentration of DNA is then checked with NanoDrop.
Materials and Protocol
Please refer to:
Results
The bands we got from gel electrophoresis is very faint.
Conclusion
We realized that we used the wrong rocF primer, so we repeat the whole procedure from PCR again.
rocF and Subtilin immunity
Gel extraction
Aims
We digested the plasmid pSB1C3 with the restriction enzyme HindIII to linearize it and then we would be running the sequence on the gel to check if the plasmid has become linear or not and then we would be extracting it.
Materials and protocol
Please refer to the:
- gel electrophoresis,
- gel extraction and
- NanoDrop spectrophotometer protocols.
Results
- Lane 1: 1 Kb ladder
- Lane 2: Linearized plasmid pSB1C3
- Lane 3: 1 Kb ladder
There is no gel photograph because we want to keep the exposure of DNA to the UV light to an absolute minimum.
Discussion
During gel extraction procedure, we found a bright band of approx During gel extraction procedure, we found a bright band of approximately 3100 bp size in lane 2 under UV light and we cut the gel and extracted the band.
Conclusion
We got linearized plasmid pSB1C3 and we performed gel extraction successfully and the nanodrop protocol showed that we got 12.7 ng/µl concentration of plasmid.
PCR
Aim
The aim of this experiment is to amplify plasmid linearized pSB1C3 at 3 different melting temperatures for the construction of rocF BioBrick with the help of 3 different Phusion PCR.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol. The details for the 3 PCR reactions are mentioned below:
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | pSB1C3 | pSB1C3_for | pSB1C3_rev | 64 | 2072 approx. | 65 |
| 2 | Plasmid Vector | pSB1C3 | pSB1C3_for | pSB1C3_rev | 65 | 2072 approx. | 65 |
| 3 | Plasmid Vector | pSB1C3 | pSB1C3_for | pSB1C3_rev | 66 | 2072 approx. | 65 |
Table 2: Table represents 3 different Phusion PCR reactions for the amplification of linearized plasmid pSB1C3, Pspac_oid promoter and double treminator so that it can be ligated together with other fragments for the construction of rocF with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different.
- For more about the rocF fragments, please refer to the Cloning strategy for rocF.
Discussion
All the 3 Phusion PCR reactions were done however, gel electrophoresis will be done tomorrow, to check whether the fragments have actually amplified or not.
Conclusion
Tomorrow we will be running gel electrophoresis to check the outcome of the 3 PCR reactions and later all the fragments will be ligated with help of Gibson protocol if the fragments have amplified.
 
|
 "
"