Team:Newcastle/24 August 2010
From 2010.igem.org
(→Results) |
Shethharsh08 (Talk | contribs) (→Results) |
||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | =Miniprep for pGFPrrnB with | + | =Miniprep for pGFPrrnB with filamentous cell part = |
==Aims== | ==Aims== | ||
| - | The aim of this experiment is to | + | The aim of this experiment is to obtain stock of the filamentous cell part in pGFP-rrnB for transformation into ''Bacillus subtilis'' 168. |
==Materials and Protocol== | ==Materials and Protocol== | ||
| Line 48: | Line 48: | ||
The concentration ranged from 286.1 µl/ml to 518.7 µl/ml. Therefore we have obtained high concentration of ''yneA'' in pGFPrrnB. | The concentration ranged from 286.1 µl/ml to 518.7 µl/ml. Therefore we have obtained high concentration of ''yneA'' in pGFPrrnB. | ||
| - | = | + | =Double Digestion of pGFPrrnB with filamentous cell part= |
==Aims== | ==Aims== | ||
| - | The aim of this experiment is to check if the insert | + | The aim of this experiment is screen our minipreps to check if the insert is present. |
==Materials and Protocol== | ==Materials and Protocol== | ||
| Line 58: | Line 58: | ||
We are doing two digests for pGFPrrnB and ''yneA'': | We are doing two digests for pGFPrrnB and ''yneA'': | ||
| - | * Single digest with | + | * Single digest with HindIII; |
| - | * Double digest with | + | * Double digest with EcoRI and NheI. |
Please refer to [[Team:Newcastle/Restriction_digests|restriction digests]] and [[Team:Newcastle/Gel_electrophoresis|gel electrophoresis]]. | Please refer to [[Team:Newcastle/Restriction_digests|restriction digests]] and [[Team:Newcastle/Gel_electrophoresis|gel electrophoresis]]. | ||
| - | == | + | ==Results== |
| - | |||
| - | + | [[Image:Picture5.1.png|400px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Gel electrophoresis | + | '''Figure 1''': Gel electrophoresis result for restriction digest of pGFPrrnB and ''yneA'' with Nhe1 and Spe1. |
| - | + | * '''Lane 1''': 1kb DNA ladder | |
| + | * '''Lane 2''': Tube 1 | ||
| + | * '''Lane 3''': Tube 2 | ||
| + | * '''Lane 4''': Tube 3 | ||
| + | * '''Lane 5''': Tube 4 | ||
| + | * '''Lane 6''': Tube 5 | ||
| + | * '''Lane 7''': Tube 6 | ||
| + | * '''Lane 8''': Tube 7 | ||
| + | * '''Lane 9''': Tube 8 | ||
| + | * '''Lane 10''': Tube 9 | ||
| + | * '''Lane 11''': Tube 10 | ||
| + | * '''Lane 12''': Tube 11 | ||
| + | * '''Lane 13''': Tube 12 | ||
| + | * '''Lane 14''': 1kb DNA ladder | ||
| - | + | ==Conclusion== | |
| - | + | The results show that the insert is present, as there is a band at approximately 541bp corresponding to ''yneA'' and a band at approximately 8.4kbp corresponding to pGFPrrnb in lanes 2, 3, 4, 6, 7, 8, 9, 11 and 12. | |
| - | + | ||
| - | |||
| - | |||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 02:23, 28 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Miniprep for pGFPrrnB with filamentous cell part
Aims
The aim of this experiment is to obtain stock of the filamentous cell part in pGFP-rrnB for transformation into Bacillus subtilis 168.
Materials and Protocol
Please refer to qiagen minipreps and nanodrop spectrophotometer protocols.
Results
| Tube 1 | Tube 2 | Tube 3 | Tube 4 | Tube 5 | Tube 6 | Tube 7 | Tube 8 | Tube 9 | Tube 10 | Tube 11 | Tube 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 286.1 µl/ml | 304.0 µl/ml | 316.3 µl/ml | 421.6 µl/ml | 518.7 µl/ml | 460.1 µl/ml | 370.1 µl/ml | 377.3 µl/ml | 346.0 µl/ml | 347.4 µl/ml | 202.8 µl/ml | 307.4 µl/ml |
Table 1: Nanodrop spectrophotometer results. Table represents the amount of plasmid present in µl/ml quantity.
Discussion
The concentration ranged from 286.1 µl/ml to 518.7 µl/ml. Therefore we have obtained high concentration of yneA in pGFPrrnB.
Double Digestion of pGFPrrnB with filamentous cell part
Aims
The aim of this experiment is screen our minipreps to check if the insert is present.
Materials and Protocol
We are doing two digests for pGFPrrnB and yneA:
- Single digest with HindIII;
- Double digest with EcoRI and NheI.
Please refer to restriction digests and gel electrophoresis.
Results
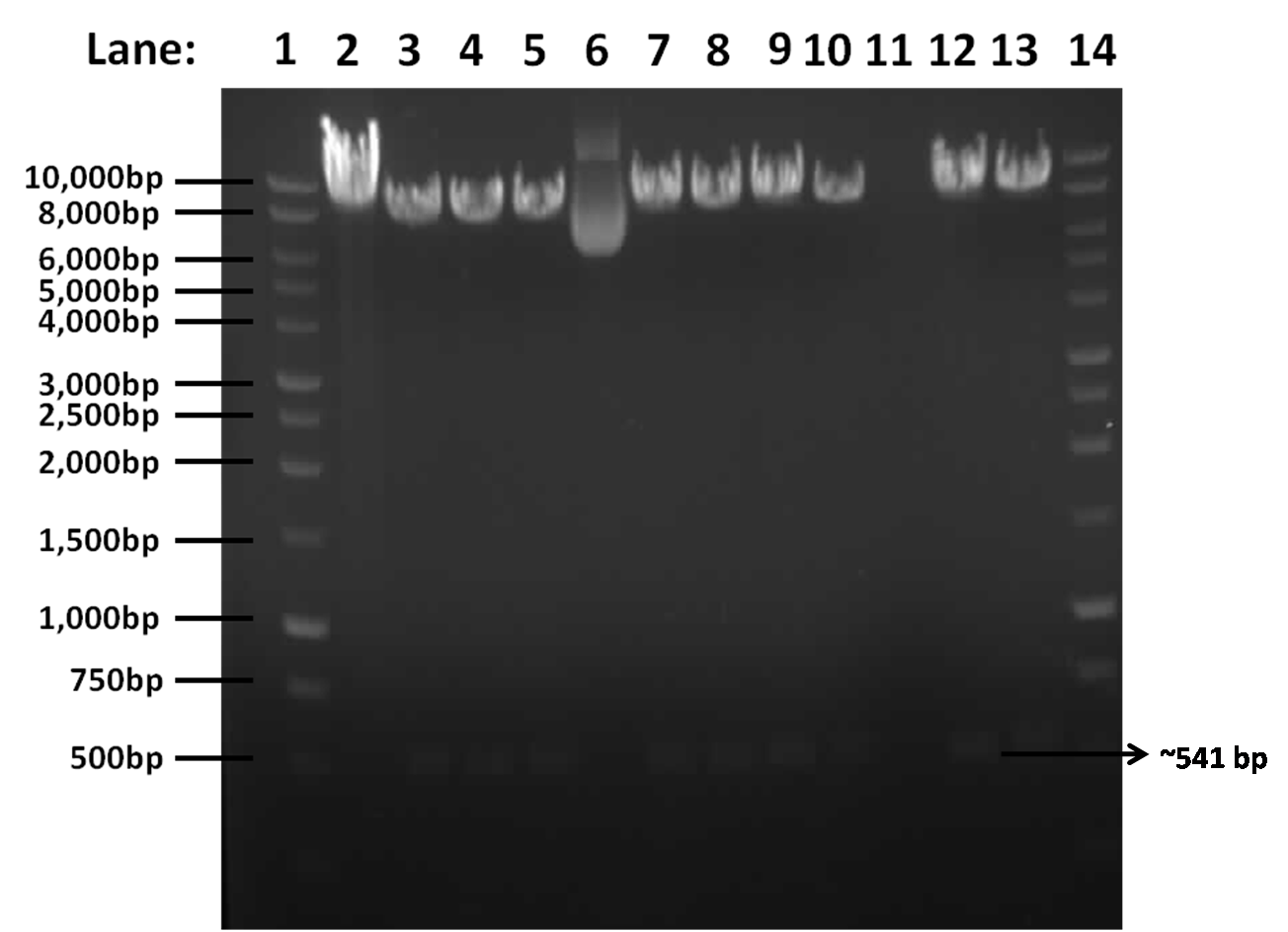
Figure 1: Gel electrophoresis result for restriction digest of pGFPrrnB and yneA with Nhe1 and Spe1.
- Lane 1: 1kb DNA ladder
- Lane 2: Tube 1
- Lane 3: Tube 2
- Lane 4: Tube 3
- Lane 5: Tube 4
- Lane 6: Tube 5
- Lane 7: Tube 6
- Lane 8: Tube 7
- Lane 9: Tube 8
- Lane 10: Tube 9
- Lane 11: Tube 10
- Lane 12: Tube 11
- Lane 13: Tube 12
- Lane 14: 1kb DNA ladder
Conclusion
The results show that the insert is present, as there is a band at approximately 541bp corresponding to yneA and a band at approximately 8.4kbp corresponding to pGFPrrnb in lanes 2, 3, 4, 6, 7, 8, 9, 11 and 12.
 
|
 "
"