Team:Newcastle/23 August 2010
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
pSB1C3 plasmid gel electrophoresis
Aims
The aim of this experiment is to run gel electrophoresis for the extracted and linearized plasmid pSB1C3 fragment which were amplified at 4 different melting temperatures by 4 separate PCR reactions.
Materials and Protocol
Please refer to: gel electrophoresis for gel electrophoresis protocol.
Result
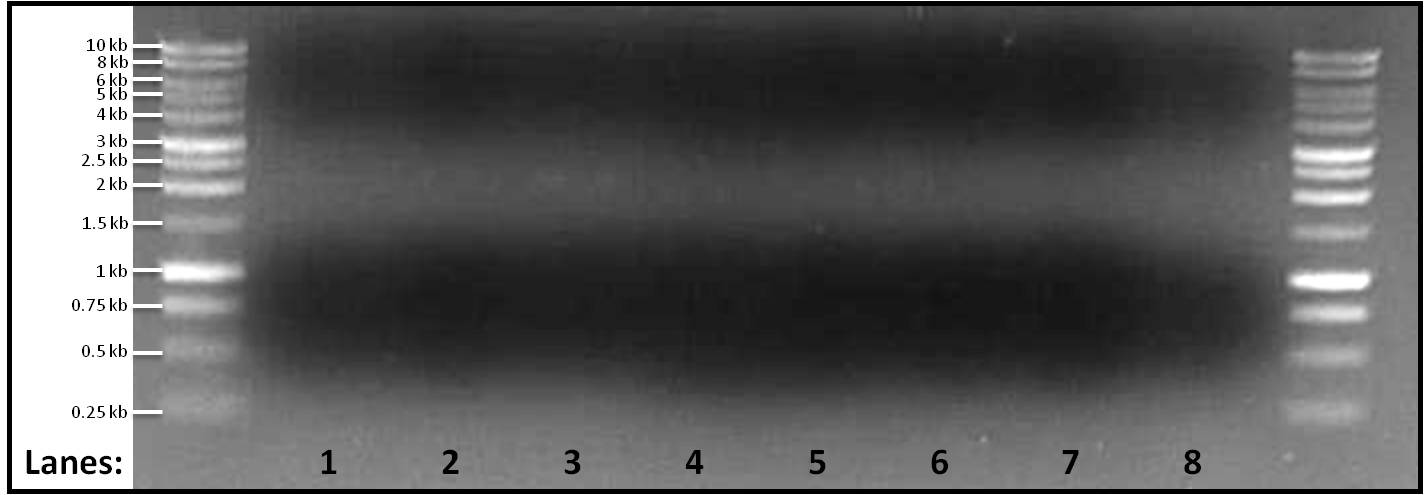
Figure 1: Gel electrophoresis of the amplified linearized plasmid pSB1C3 fragments ran at 4 different melting temperatures, Tms, (50, 60, 65, 70°C). A 1 kb DNA ladder was used on either side of lanes.
- Lane 1: pSB1C3 fragment amplified at 55°C
- Lane 2: pSB1C3 fragment amplified at 60°C
- Lane 3: pSB1C3 fragment amplified at 65°C
- Lane 4: pSB1C3 fragment amplified at 70°C
- Lane 5: pSB1C3 fragment amplified at 55°C
- Lane 6: pSB1C3 fragment amplified at 60°C
- Lane 7: pSB1C3 fragment amplified at 65°C
- Lane 8: pSB1C3 fragment amplified at 70°C
Discussion
No bands were found in any of the lanes. Yesterday, a faint band was found when the melting temperature was set at 65°C but today no band is found in lane 3 and lane 7. This makes finding the cause for no amplification even difficult. We would still be looking into it and would be changing other parameters.
Conclusion
The PCR reaction failed as there is no amplification found in any of the reactions.
pSB1C3 plasmid gel electrophoresis by adding EtBr
Aims
The aim of this experiment is to run gel electrophoresis for the extracted and linearized plasmid pSB1C3 fragment which were amplified at 4 different melting temperatures by 4 separate PCR reactions and to put Ethidium Bromoide (EtBr) in the agarose gel instead of safeview die so as to get better resolution and brighter bands.
Materials and Protocol
Please refer to: gel electrophoresis for gel electrophoresis protocol.
Please note that in this protocol we have made a minor modification which is that instead of adding 5µl of safeview dye, we have added 5µl of EtBr into the agarose gel so as to get better resolution and brighter bands.
Result
Figure 2: Gel electrophoresis (containing EtBr) of the amplified linearized plasmid pSB1C3 fragments ran at 4 different melting temperatures, Tms, (50, 60, 65, 70°C). A 1 kb DNA ladder was used on either side of lanes.
- Lane 1: pSB1C3 fragment amplified at 55°C
- Lane 2: pSB1C3 fragment amplified at 60°C
- Lane 3: pSB1C3 fragment amplified at 65°C
- Lane 4: pSB1C3 fragment amplified at 70°C
- Lane 5: pSB1C3 fragment amplified at 55°C
- Lane 6: pSB1C3 fragment amplified at 60°C
- Lane 7: pSB1C3 fragment amplified at 65°C
- Lane 8: pSB1C3 fragment amplified at 70°C
Discussion
No bands were found in any of the lanes. Yesterday, a faint band was found when the melting temperature was set at 65°C but today no band is found in lane 3 and lane 7. This makes finding the cause for no amplification even difficult. We would still be looking into it and would be changing other parameters.
Conclusion
The PCR reaction failed as there is no amplification found in any of the reactions.
pSB1C3 plasmid gel electrophoresis by modifying PCR consitions
Aims
The aim of this experiment is to run gel electrophoresis for the extracted and linearized plasmid pSB1C3 fragment which were amplified at 4 different melting temperatures by 4 separate PCR reactions and to put Ethidium Bromoide (EtBr) in the agarose gel instead of safeview die so as to get better resolution and brighter bands.
Materials and Protocol
Please refer to: gel electrophoresis for gel electrophoresis protocol.
Please note that in this protocol we have made a minor modification which is that instead of adding 5µl of safeview dye, we have added 5µl of EtBr into the agarose gel so as to get better resolution and brighter bands.
Result
Figure 2: Gel electrophoresis (containing EtBr) of the amplified linearized plasmid pSB1C3 fragments ran at 4 different melting temperatures, Tms, (50, 60, 65, 70°C). A 1 kb DNA ladder was used on either side of lanes.
- Lane 1: pSB1C3 fragment amplified at 55°C
- Lane 2: pSB1C3 fragment amplified at 60°C
- Lane 3: pSB1C3 fragment amplified at 65°C
- Lane 4: pSB1C3 fragment amplified at 70°C
- Lane 5: pSB1C3 fragment amplified at 55°C
- Lane 6: pSB1C3 fragment amplified at 60°C
- Lane 7: pSB1C3 fragment amplified at 65°C
- Lane 8: pSB1C3 fragment amplified at 70°C
Discussion
No bands were found in any of the lanes. Yesterday, a faint band was found when the melting temperature was set at 65°C but today no band is found in lane 3 and lane 7. This makes finding the cause for no amplification even difficult. We would still be looking into it and would be changing other parameters.
Conclusion
The PCR reaction failed as there is no amplification found in any of the reactions.
 
|
 "
"