Team:Newcastle/22 June 2010
From 2010.igem.org
RachelBoyd (Talk | contribs) (→Set up ligation) |
Shethharsh08 (Talk | contribs) (→Nanodrop Protocol) |
||
| (20 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | == | + | ==Aims== |
| + | The aim of today's Lab practice was to extract the plasmids for GFP and RFP, digest the 2 plasmids and extract the 2 inserts and 1 of the backbones (vector)from an agarose gel. From this a ligation was set up. | ||
| - | + | ==Materials and Protocol== | |
| - | + | ||
| - | + | ||
| - | === | + | ===QIagen Miniprep Kit Using a Microcentrifuge for Plasmid Extraction=== |
| + | Please refer to: [[Team:Newcastle/Qiagen Minipreps|Qiagen Minipreps]] for materials required and protocol. | ||
| - | === | + | ===Digest=== |
| + | Please refer to: [[Team:Newcastle/Restriction digests| Restriction digests]] for materials required and protocol. | ||
| - | + | ===Gel extraction=== | |
| - | + | Please refer to: [[Team:Newcastle/Gel extraction| Gel extraction]] for materials required and protocol. | |
| - | === | + | |
| + | ===Agarose Gel Electrophoresis=== | ||
| + | Please refer to: [[Team:Newcastle/Gel electrophoresis| Gel electrophoresis]] for materials required and protocol. | ||
| - | |||
{| | {| | ||
|- | |- | ||
| - | |[[Image:Newcastle_lab_1.jpeg|thumb]] | + | |[[Image:Newcastle_lab_1.jpeg|thumb|Dr Wendy demostrating how to use gel electrophoresis machine]] |
| - | |[[Image:Newcastle_lab_2.jpeg|thumb]] | + | |[[Image:Newcastle_lab_2.jpeg|thumb|Gel tank]] |
| - | |[[Image:Newcastle_lab_3.jpeg|thumb]] | + | |[[Image:Newcastle_lab_3.jpeg|thumb|Deena pouring hot molten gel]] |
|- | |- | ||
|} | |} | ||
| Line 32: | Line 30: | ||
{| | {| | ||
|- | |- | ||
| - | |[[Image:Newcastle_lab_4.jpeg|thumb]] | + | |[[Image:Newcastle_lab_4.jpeg|thumb|Putting the gel on the geldoc]] |
| - | |[[Image:Newcastle_lab_5.jpeg|thumb]] | + | |[[Image:Newcastle_lab_5.jpeg|thumb|Preparing to cut the gel]] |
| - | |[[Image:Newcastle_lab_6.jpeg|thumb]] | + | |[[Image:Newcastle_lab_6.jpeg|thumb|Gel illuminated by the UV light which is emitted by the geldoc]] |
|- | |- | ||
|} | |} | ||
| - | We cut the inserts which were about 900bp and we use the backbone from the | + | We cut the inserts which were about 900bp and we use the backbone from the plasmid consisting ''rfp''. |
====QIAquick Gel extraction kit==== | ====QIAquick Gel extraction kit==== | ||
| Line 44: | Line 42: | ||
{| | {| | ||
|- | |- | ||
| - | |[[Image:Newcastle_lab_7.jpeg|thumb]] | + | |[[Image:Newcastle_lab_7.jpeg|thumb|Phil cutting the gel]] |
| - | |[[Image:Newcastle_lab_8.jpeg|thumb]] | + | |[[Image:Newcastle_lab_8.jpeg|thumb|Transferring the cut gel part into a 2 ml microfuge tube]] |
|- | |- | ||
|} | |} | ||
| - | + | ==Set up ligation== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{|border=1 | {|border=1 | ||
| Line 99: | Line 91: | ||
|10.0 | |10.0 | ||
|} | |} | ||
| + | ==Transformation protocol== | ||
| + | Please refer to: [[Team:Newcastle/Transformation of E. coli|Transformation of ''E. coli'']] for materials required and protocol. | ||
| + | |||
| + | ==Nanodrop Protocol== | ||
| + | Please refer to: [[TeamNewcastleNanoDrop Spectrophotometer| Nanodrop Spectrophotometer]] for materials required and protocol. | ||
| - | |||
{| | {| | ||
|- | |- | ||
| - | |[[Image:Newcastle_nanodrop_1.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_1.jpeg|thumb|A typical curve to be achieved from a nanodrop spectrophotmeter]] |
| - | |[[Image:Newcastle_nanodrop_2.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_2.jpeg|thumb|Phil analyzing a sample on the nanodrop machine]] |
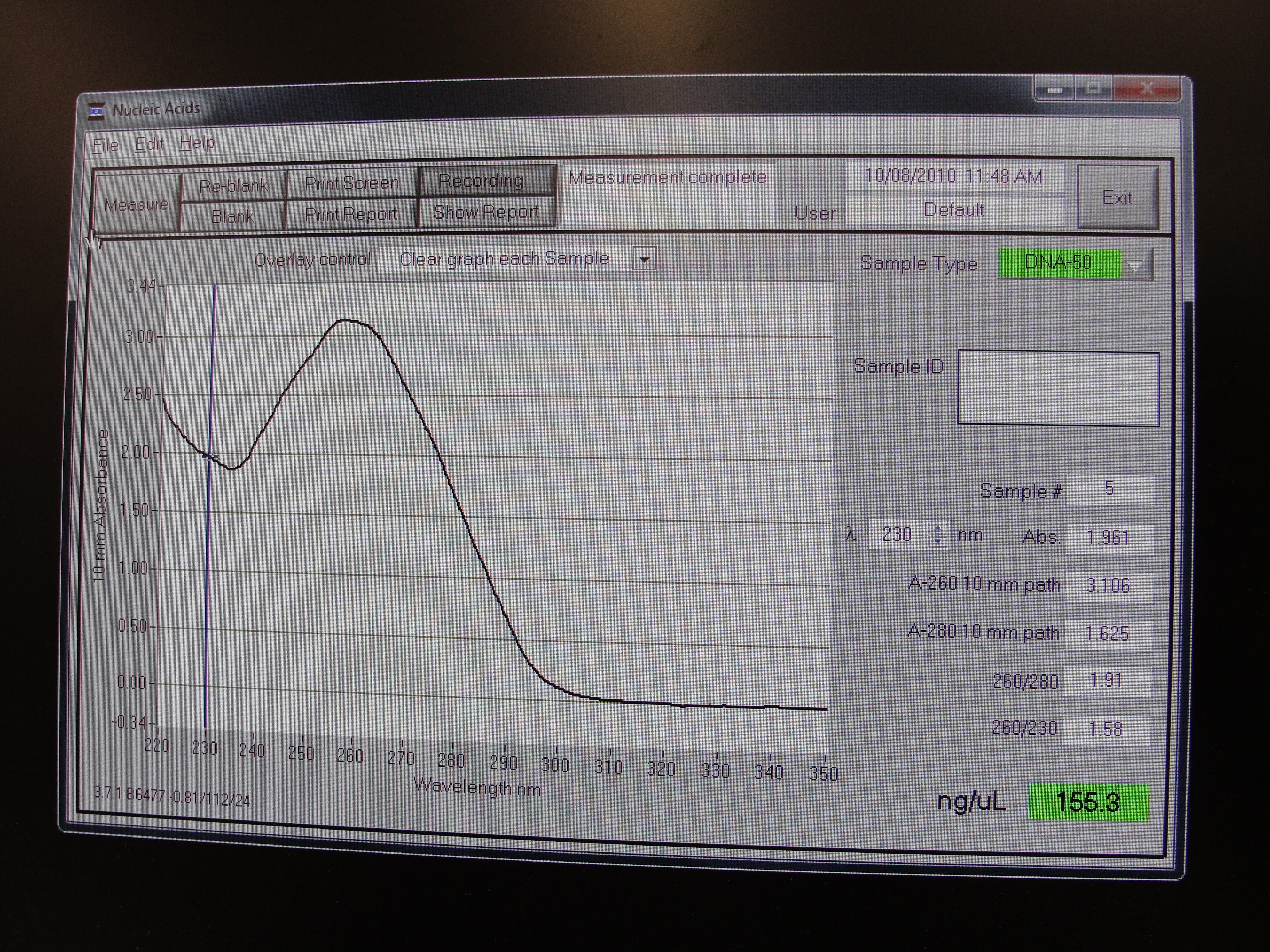
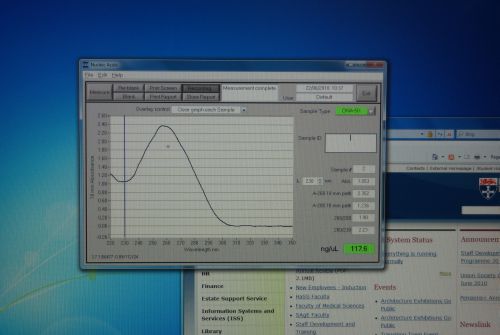
| - | |[[Image:Newcastle_nanodrop_3.jpeg|thumb]] | + | |[[Image:Newcastle_nanodrop_3.jpeg|thumb|Analyzed data on the screen]] |
|- | |- | ||
| + | |} | ||
| + | |||
| + | ==Outcome== | ||
| + | |||
| + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="2" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Team:Newcastle|Home]] | ||
| + | !align="center"|[[Team:Newcastle/Team|Team]] | ||
| + | !align="center"|[https://igem.org/Team.cgi?year=2010&team_name=Newcastle Official Team Profile] | ||
| + | !align="center"|[[Team:Newcastle/Project|Project]] | ||
| + | !align="center"|[[Team:Newcastle/Parts|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Newcastle/Modelling|Modelling]] | ||
| + | !align="center"|[[Team:Newcastle/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Newcastle/Safety|Safety]] | ||
|} | |} | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 20:42, 21 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Aims
The aim of today's Lab practice was to extract the plasmids for GFP and RFP, digest the 2 plasmids and extract the 2 inserts and 1 of the backbones (vector)from an agarose gel. From this a ligation was set up.
Materials and Protocol
QIagen Miniprep Kit Using a Microcentrifuge for Plasmid Extraction
Please refer to: Qiagen Minipreps for materials required and protocol.
Digest
Please refer to: Restriction digests for materials required and protocol.
Gel extraction
Please refer to: Gel extraction for materials required and protocol.
Agarose Gel Electrophoresis
Please refer to: Gel electrophoresis for materials required and protocol.
Cut gel out
We cut the inserts which were about 900bp and we use the backbone from the plasmid consisting rfp.
QIAquick Gel extraction kit
Set up ligation
| Reagents | 1:3(μl) | 1:5(μl) | Vector(μl) |
|---|---|---|---|
| V | 0.8 | 0.8 | 1 |
| G | 2.7 | 4 | |
| R | 5.4 | 7.7 | |
| LT4 | 1 | 1 | 1 |
| LB | 1.1 | 1.5 | 1 |
| H2O | 0 | 0 | 7 |
| Total Volume | 11.0 | 15.0 | 10.0 |
Transformation protocol
Please refer to: Transformation of E. coli for materials required and protocol.
Nanodrop Protocol
Please refer to: Nanodrop Spectrophotometer for materials required and protocol.
Outcome
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modelling | Notebook | Safety |
|---|
 
|
 "
"