Team:Newcastle/19 August 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) (→Aims) |
Shethharsh08 (Talk | contribs) (→Materials and protocol) |
||
| Line 204: | Line 204: | ||
Please refer to the: | Please refer to the: | ||
| - | *[[Team:Newcastle/Gel_electrophoresis| gel electrophoresis]], | + | *[[Team:Newcastle/Gel_electrophoresis| gel electrophoresis]], |
*[[Team:Newcastle/Gel_extraction| gel extraction]] and | *[[Team:Newcastle/Gel_extraction| gel extraction]] and | ||
*[[TeamNewcastleNanoDrop_Spectrophotometer| NanoDrop spectrophotometer]] protocols. | *[[TeamNewcastleNanoDrop_Spectrophotometer| NanoDrop spectrophotometer]] protocols. | ||
Revision as of 09:56, 26 August 2010

| |||||||||||||
| |||||||||||||
Contents |
yneA
Gel extraction
Aims
To purify the products of yneA, pGFPrrnB and PSB1AT3 by extracting the bands from the gel after running gel electrophoresis. Concentration of the DNA and vectors are then checked with nanodrop.
Materials and Protocol
Please refer to
Results
| yneA 1 | yneA 2 | yneA 3 | pSB1C3 | pGFPrrnB | |
|---|---|---|---|---|---|
| Concentration of DNA ng/µl | 3.4 | 4.7 | 5.4 | 2.2 | 18.5 |
Discussion
The bands we extracted from the gel was good for pGFPrrnB, but not so strong for yneA and PSB1C3.
The results we got from the nanodrop were not as good as expected for yneA and PSB1C3.
Conclusion
We will proceed with digestion, but another set of ligation will be set up to repeat the process again.
Ligation
Aims
To ligate yneA into both vectors pGFPrrnB and pSB1C3.
Materials and Protocol
Please refer to ligation.
Ligation mix for pSB1C3 with yneA
The concentration of pSB1C3 and yneA that we got from the gel extraction earlier today was very low, so we used a different mixture set up for ligation.
Ligation mix:
| Reagents | 1:3(μl) | 1:5(μl) | Vector(μl) |
|---|---|---|---|
| Vector | 3 | 2 | 1.5 |
| Insert | 3 | 6 | 7.5 |
| 10X BUFFER | 1 | 1 | 1.1 |
| T4 Ligase | 1 | 1 | 1 |
| H2O | 2 | 0 | 0 |
| Total Volume | 10.0 | 10.0 | 10.0 |
Results and Conclusion
We will leave the ligation overnight because concentration of yneA, pGFPrrnB and pSB1C3 are not very good. Please refer to 20.08.10 for results and discussion.
Digestion
Aim
To repeat digestion from yesterday.
Materials and Protocol
Please refer to restriction digest.
Setting up Overnight Cultures
Aims
To prepare cultures for miniprep of yneA, pGFPrrnB and pSB1C3 tomorrow.
Materials and Protocol
Please refer to growing an overnight culture.
Subtilin Immunity
Aims
Today we received the four new primers that we ordered:
- Prom_For
- pSB1C3_for
- pSB1C3_rev
- Term_rev
These primers were ordered as a solution to the problem we encountered before we were about to attempt carry out the Gibson protocol for the Subtilin Immunity (and RocF) BioBrick. The primers will be used with previously rehydrated primers to try and obtain all four parts that we need to make the Subtilin Immunity BioBrick using the Gibson Protocol. So far the spaIFEG coding sequence (part 3) has already been successfully obtained. We still need to obtain the plasmid vector (part 1), promoter & RBS (part 2) and double terminator (part 4). So in order to do this the primers received today will be rehydrated and Phusion PCR will be carried out for each of the parts in order to amplify these parts.
Materials and protocol
Please refer to DNA rehydration page for the protocol for the rehydration of the four primers and to the Phusion PCR page for protocol followed for Phusion PCR. Table # below shows the different fragments, primers and conditions used for each tube.
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | pSB1C3 | pSB1C3_for | pSB1C3_rev | 68 | 2046 + | 70 |
| 2 | Promoter and RBS (pVeg-SpoVG) | BioBrick Bba_K143053 | Prom_for | P2P2_rev | 67 | 139 + | 15 |
| 4 | Double terminator | pSB1AK3 consisting BBa_B0014 | P1T1_for | Term_rev | 63 | 116 + | 15 |
Table 1: This table shows the three different Phusion PCR reactions that were carried out today. If this is successful, these three parts (part 1, 2 and 4) along with (part 3) which is already obtained can be ligated together for the construction of subtilin immunity BioBrick with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1 Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different.
- The primers in bold are the new primers we received today. The primers not in bold were previously re-hydrated.
Discussion and Conclusion
Tommorrow we will carry out a test gel to check the sizes of the amplified fragments and if the sizes are correct then gel extraction will be performed
rocF BioBrick
Gel extraction of linearised pSB1C3
Aims
We digested the plasmid pSB1C3 with the restriction enzyme EcoR1 to linearize it and then we would be running the sequence on the gel to check if the plasmid has become linear or not and then we would be extracting it.
Materials and protocol
Please refer to the:
- gel electrophoresis,
- gel extraction and
- NanoDrop spectrophotometer protocols.
Results
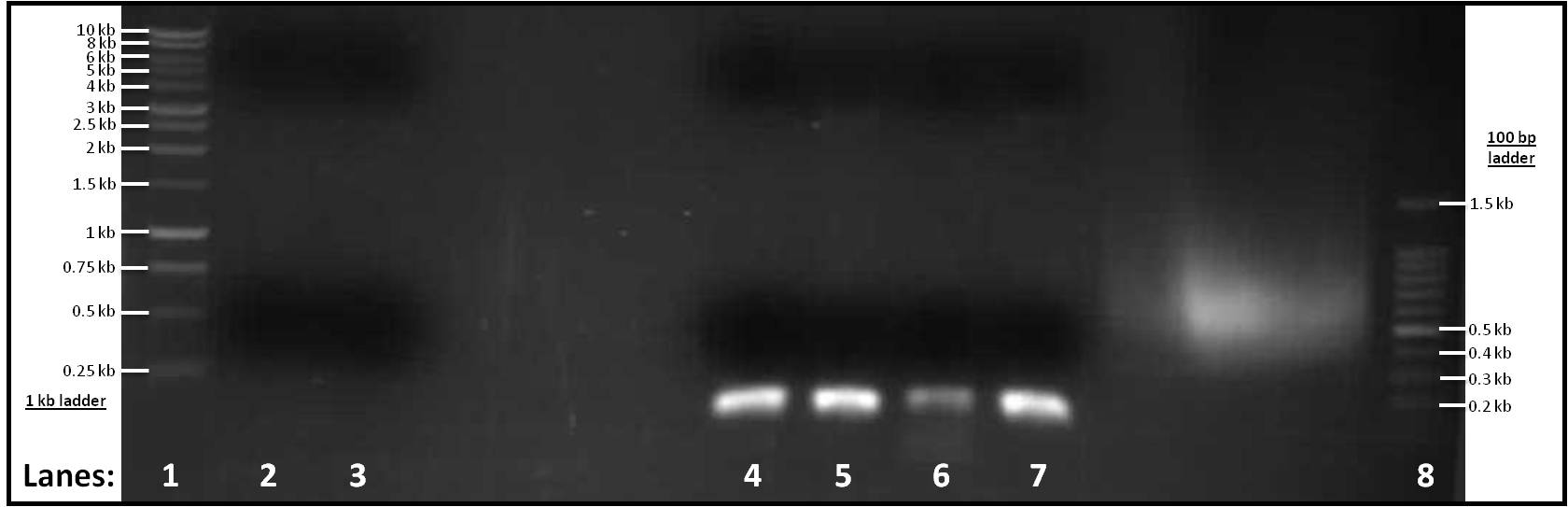
Figure 1: Gel electrophoresis of the amplified PCR products
- Lane 1: 1 Kb ladder
- Lane 2: pSB1C3 (for Subtilin Immunity)
- Lane 3: pSB1C3 (for rocF)
- Lane 4: pVeg (for Subtilin Immunity)
- Lane 5: terminator (for Subtilin Immunity)
- Lane 6: pSpacoid (for rocF)
- Lane 7: terminator (for rocF)
- Lane 8: 100 bp ladder
Discussion
After looking at the results of our gel, there are no distinct band in lanes 2-3, therefore pSB1C3 vector did not work for both Subtilin Immunity or rocF. However, there are single bands in lanes 4-7 at similar positions to each other. They are about ... in size which is what is expected.
As lanes 2-3 showed no bands, we decided to make a range of Tms to perform another set of PCR reactions to check whether the Tm was the problem. Three Tms were used: 60°C, 65°C and 70°C and six tubes were set up; a duplicate was created for each Tm. After the PCR reactions were finished, there still wasn't any visible pSBIC3 vector band showing up. The problem could have occurred due to the working stocking solution for our primers. Our next step was to re-make the working stock solution for our primers. Also, we had decided to use the Tms 55°C, 60°C, 65°C, 70°C - duplicate for each, making eight PCR tubes in total.
As lanes 4-7 had bands at the right positions, gel extraction of these four parts was carried out. The concentration of DNA was then measured using the Nanodrop spectrophotometer.
Conclusion
The PCR machines were left to run and results of the eight PCR reactions at the 4 different temperatures shall be obtained on Monday. After a test gel has been run and if bands appear at the right sizes then gel extraction will be carried out.
PCR of pSB1C3, pspac oid and double terminator with new primers
Go back to our main Lab book page
 
|
 "
"