Team:Nevada/Agrobacterium Transformations
From 2010.igem.org
m (→Agrobacterium Transformations as an Alternative to NT Cell Transformations) |
|||
| Line 7: | Line 7: | ||
<p> </p> | <p> </p> | ||
| - | == Agrobacterium | + | == How Agrobacterium Transforms Plants == |
<html> | <html> | ||
Revision as of 20:14, 26 October 2010
How Agrobacterium Transforms Plants
Agrobacterium Transformation of Plants
Agrobacterium mediated transformation is the most widely used means of integrating DNA fragments of interested into the genome of plant cells. This process takes advantage of the naturally occurring plant pathogen, Agrobacterium tumefaciens. This soil bacterium is the causative agent responsible for crown gall disease in a wide range of plant species. Pathogenic Agrobacterium carry a large plasmid, referred to as the Tumor Inducing or Ti plasmid, which is required for disease transmission. The Ti plasmid contains virulence genes (vir) which code for the protein machinery required for transfer and integration of disease causing genes into the host plants genome. The Ti plasmid also contains a set of genes designated as the T-DNA. The T-DNA is flanked by inverted repeat sequences referred to as the Left Border (LB) and Right Border (RB) sequences. Any DNA sequences situated between the LB and RB will be inserted into the plant host genome. In pathogenic strains of A. tumefaciens, the border sequences flank genes involved in tumor formation in infected plants. Once integrated into the host genome, these genes, referred to as the oncogenes, hijack the host plant to produce the protein machinery needed for the production of phytohormones. These phytohormones cause uncontrolled cell proliferation which resulting in the formation of a gall or tumor. In addition to the oncogenes, the T-DNA region also encoded biosynthetic proteins for the production of unusual amino acids referred to as opines. Therefore, by integrating the T-DNA into the host, the bacteria forces the plant host to create both a habitat and a carbon/nitrogen source on which the bacteria could survive.
Development of Agrobacterium as a Vector for Plant Genetic Engineering
In the 1980’s scientist took advantage of A. tumefacien’s ability to insert foreign DNA into plant chromosomes to develop highly efficient vectors for plant transformation and genetic engineering. By swapping the oncogenes from the T-DNA region of the Ti plasmid with any plant gene expression cassette (containing a plant promoter, a gene of interest and a transcriptional terminator sequence), they were able to integrate their particular gene of interest into the host genome without causing the disease. They later found that the vir gene products could function in trans such that the T-DNA present on a separate plasmid from the plasmid carrying the vir genes could still be transferred to the host plant. This system referred to as the binary transformation system was composed of an Agrobacterium strain containing a Ti plasmid in which the T-DNA was deleted and a second plasmid which carries the LB and RB sequences flanking two plant gene expression cassettes. One of these cassettes was used to express a gene of interest and the second was used to express an antibiotic resistance gene for the selection of positive transformation events.
Molecular Progression of Agrobacterium Infection
1. chemical perception of host
2. induction of virulence genes
3. formation of infection apparatus
4. preparation of T-DNA
5. export T-DNA to plant cell
6. import T-DNA to plant nucleus
7. integration of T-DNA into host genome
8. expression of T-DNA
Agrobacterium Transformation "Up Close":

Agrobacterium Transformation Overview:
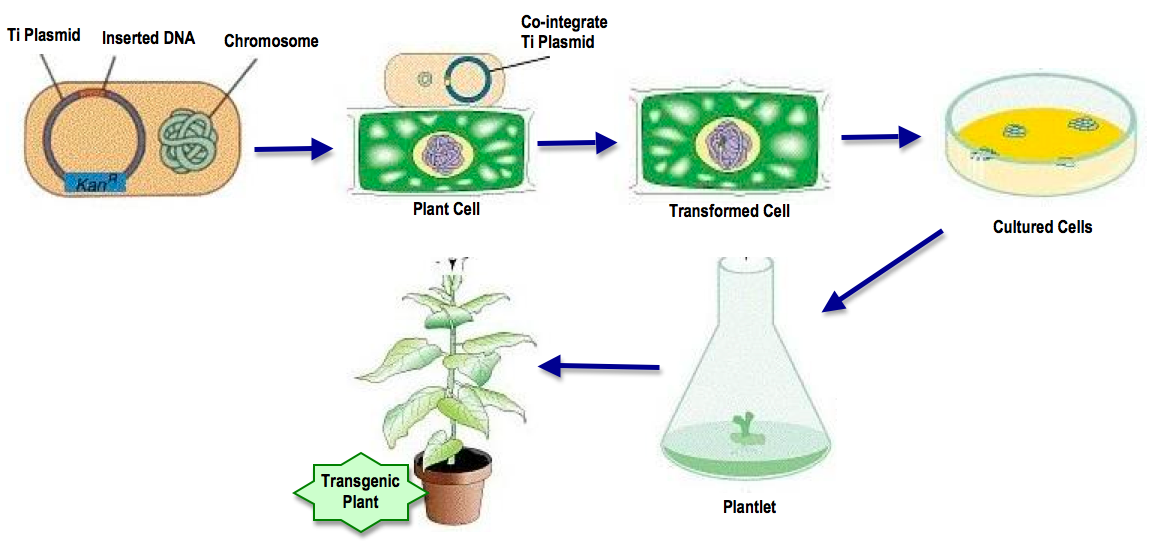
 "
"