Team:NYU/Project
From 2010.igem.org
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | < | + | <html> |
| - | + | <div id="allcontent"> | |
| - | + | <img class="bannerimage" src="https://static.igem.org/mediawiki/2010/2/2d/NYU_bannerv_6.png" height="200px" width="950px"><br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <div id="headerlinks"> | |
| - | + | <ul id="nav"> | |
| - | + | <li> | |
| - | + | <a class="bannertoplinks" href="https://2010.igem.org/Team:NYU">Home</a> | |
| - | + | </li> | |
| - | + | ||
| - | + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Project">Project</a> | |
| - | + | <ul style="z-index:1"> | |
| - | + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Project">Overview</a></li> | |
| - | + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Experiments">Experiments</a></li> | |
| - | + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Assembly">Overlap Assembly</a></li> | |
| - | + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Notebook">Notebook</a></li> | |
| - | + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Parts">Biobricks</a></li> | |
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Programming">Programming</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Team">Team</a> | ||
| + | <ul style="z-index:1"> | ||
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Team">NYU</a></li> | ||
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/CornellMed">Cornell Med</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a> | ||
| + | <ul style="z-index:1"> | ||
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a></li> | ||
| + | <li><a class="bannerlinks" href="http://www.sciencehouse.com">ScienceHouse</a></li> | ||
| + | <li><a class="bannerlinks" href="http://www.nysynbio.org">NY synbio</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Contact">Contact Us</a> | ||
| + | |||
| + | </li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </p><p><br /> | ||
| + | |||
| + | <style type="text/css"> | ||
| + | |||
| + | |||
| + | #allcontent { | ||
| + | width: 950px; | ||
| + | } | ||
| + | |||
| + | |||
| + | body { | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | font-family: Helvetica, Arial, sans-serif; | ||
| + | width: 900px; | ||
| + | } | ||
| + | |||
| + | table { | ||
| + | color: #000000; | ||
| + | font-family: Helvetica, Arial, sans-serif; | ||
| + | } | ||
| + | |||
| + | #content { | ||
| + | color: #000000; | ||
| + | font-family: Helvetica, Arial, sans-serif; | ||
| + | width: 965px; | ||
| + | line-height: 1.2em; | ||
| + | |||
| + | } | ||
| + | |||
| + | a { | ||
| + | text-decoration: none; | ||
| + | color: #7F5217; | ||
| + | } | ||
| + | |||
| + | a:hover { | ||
| + | color: #A0C544; | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | h1, h2, h3, h4, h5, h6 { | ||
| + | color: #254117; | ||
| + | } | ||
| + | |||
| + | #headerlinks{ | ||
| + | margin-left: -20px; | ||
| + | } | ||
| + | |||
| + | #nav, #nav ul { | ||
| + | padding: 0; | ||
| + | list-style: none; | ||
| + | } | ||
| + | |||
| + | #nav a { | ||
| + | display: block; | ||
| + | width: 149px; | ||
| + | } | ||
| + | |||
| + | #nav li { | ||
| + | float: left; | ||
| + | width: 149px; | ||
| + | margin-right: 41px; | ||
| + | } | ||
| + | |||
| + | #nav li ul { | ||
| + | position: absolute; | ||
| + | width: 149px; | ||
| + | left: -999em; | ||
| + | } | ||
| + | |||
| + | #nav li:hover ul { | ||
| + | left: auto; | ||
| + | text-decoration: none; | ||
| + | color: white !important; | ||
| + | |||
| + | } | ||
| + | |||
| + | #nav li:hover ul, #nav li.sfhover ul { | ||
| + | left: auto; | ||
| + | text-decoration: none; | ||
| + | color: white !important; | ||
| + | } | ||
| + | |||
| + | sfHover = function() { | ||
| + | var sfEls = document.getElementById("nav").getElementsByTagName("LI"); | ||
| + | for (var i=0; i<sfEls.length; i++) { | ||
| + | sfEls[i].onmouseover=function() { | ||
| + | this.className+=" sfhover"; | ||
| + | } | ||
| + | sfEls[i].onmouseout=function() { | ||
| + | this.className=this.className.replace(new RegExp(" sfhover\\b"), ""); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | if (window.attachEvent) window.attachEvent("onload", sfHover); | ||
| + | |||
| + | .bannerimage { | ||
| + | |||
| + | } | ||
| + | |||
| + | a.bannerlinks { | ||
| + | color: white !important; | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | |||
| + | a.bannertoplinks { | ||
| + | color: white !important; | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | a.bannerlinks:hover, a.bannertoplinks:hover { | ||
| + | background-color: #A0C544; | ||
| + | color: white; | ||
| + | } | ||
| + | |||
| + | .bannerlinks { | ||
| + | background-color: #698B69; | ||
| + | border-style: solid; | ||
| + | border-color: white; | ||
| + | border-width: 1px; | ||
| + | color: white; | ||
| + | padding: 3px 20px 3px 20px; | ||
| + | margin-left: -19px; | ||
| + | } | ||
| + | |||
| + | .bannertoplinks { | ||
| + | background-color: #006400; | ||
| + | border-style: solid; | ||
| + | border-color: white; | ||
| + | border-width: 1px; | ||
| + | color: white; | ||
| + | margin-bottom: -2px; | ||
| + | padding: 3px 20px 3px 20px; | ||
| + | } | ||
| + | |||
| + | #sidebar { | ||
| + | width: 300px; | ||
| + | float: right; | ||
| + | margin-right: 0px; | ||
| + | border-style: solid; | ||
| + | border-color: white; | ||
| + | border-width: 1px; | ||
| + | padding: 0px; | ||
| + | padding-top:5px; | ||
| + | } | ||
| + | |||
| + | #abstract { | ||
| + | width: 600px; | ||
| + | border-style: solid; | ||
| + | border-color: white; | ||
| + | border-width: 1px; | ||
| + | padding: 10px; | ||
| + | line-height: 1.2em; | ||
| + | font-size: 14px; | ||
| + | } | ||
| + | |||
| + | |||
| + | #rightgroup { | ||
| + | float: right; | ||
| + | width: 300px; | ||
| + | padding: 10px; | ||
| + | } | ||
| + | |||
| + | #leftgroup { | ||
| + | width: 300px; | ||
| + | padding: 10px; | ||
| + | margin-right: 20px; | ||
| + | } | ||
| + | |||
| + | #maincontent { | ||
| + | width: 610px; | ||
| + | } | ||
| + | |||
| + | #headerlinks > #nav > li { | ||
| + | border-bottom: 4px solid white; | ||
| + | } | ||
| + | |||
| + | #sidebar a{ | ||
| + | text-decoration: none; | ||
| + | color: #7F5217; | ||
| + | } | ||
| + | |||
| + | #sidebar a:hover{ | ||
| + | color: #A0C544; | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | a.sidebarlinks { | ||
| + | display: block; | ||
| + | width: 300px; | ||
| + | font-size: 1.2em; | ||
| + | text-align: center; | ||
| + | background-color: #827B60; | ||
| + | color: white !important; | ||
| + | padding: 10px 0px 10px 0px; | ||
| + | margin-bottom: 5px | ||
| + | } | ||
| + | |||
| + | a.sidebarlinks:hover { | ||
| + | background-color: #A0C544; | ||
| + | color: white !important; | ||
| + | } | ||
| + | |||
| + | a.timelinelinks { | ||
| + | color: #7F5217 !important; | ||
| + | } | ||
| + | |||
| + | a.timelinelinks:hover { | ||
| + | color: #A0C544 !important; | ||
| + | } | ||
| + | |||
| + | |||
| + | </style> | ||
| + | <!-- !END NYU CSS- Thanks Harvard --> | ||
| + | |||
| + | |||
| + | </html> | ||
| + | |||
| + | [[Image:NYU_OverviewSideF_1.png|200px|right]] | ||
| + | |||
| + | |||
| + | [[Image:NYU_logo.png|220px|left]] | ||
| + | |||
| + | |||
| + | Our goal is to use the tools of synthetic biology to make a biological machine capable of antibody discovery. To do this, we have decided to learn from our own immune system by porting its own antibody discovery strategies into an engineered strain of yeast. This yeast will be capable of screening a library of antibodies against a target antigen, recombining the antibody gene ''in vivo'' and then either secreting or surface displaying the resulting antibody protein for a variety of purposes. Our hope is to demonstrate the feasibility of using the yeast cell not only as a vessel for antibody discovery but as a streamlined processing unit that can discover and also begin production of new antibodies - all in one test tube! | ||
| Line 34: | Line 273: | ||
| - | |||
| - | + | Our plan for this year's competition is to construct an easy-to-use yeast strain capable of intracellular antibody discovery. The current dominant mode of microbial antibody discovery requires cellular surface display and high-throughput fluorescent screening. Instead of using this method, we wanted our cells to be able to sense if the antibody they are translating will bind the target antigen. | |
| - | + | [[Image:NYU_bind3.png|180px|left]] | |
| + | |||
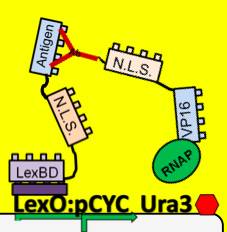
| + | Using a modified form of the yeast two-hybrid screening system, the yeast cells are able to sense the amount of antibody::antigen interaction inside the cell. When the antibody binds the target antigen it anchors the VP16 transcriptional activator near our response system. VP16 then increases the level of transcription of a reporter gene. If this gene encodes a nutritional marker, such as Ura3, only cells experiencing antibody:antigen interactions can be selected using selective media. Using this method the original population of our yeast strain will be able to, in essence, screen itself and our resulting yeast population will contain only antibodies that bind our antigen. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | In a research setting, once you have discovered the antibody you wish to use you must then reclone the gene for it into a secretion vector for production of pure antibody protein. This involves taking the plasmid out of the yeast cell, excising the antibody gene, ligating it into the second vector and then transforming it back into a suitable yeast strain. | ||
| + | |||
| + | |||
| + | [[Image:NYU_Cre.jpg|350px|left]] | ||
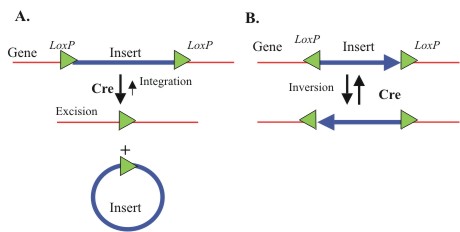
| + | To make this process faster and more easily accomplished, we have incorporated a recombination-based architecture into our system that will allow the cells to modify the antibody plasmid ''in vivo''. Using this system transcription of the antibody gene can go straight from screening to protein production in a matter of minutes. This concept has the capability to not only shave weeks off of current antibody discovery protocols, but opens the door to cellular programming for other methods, such as restructuring of antibody genes or performing more complex screenings for different purposes. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == The Experiments == | ||
| + | In the spectrum of molecular biology, iGEM is a very short time frame. Because of this short time frame, our wet lab team having only two members and our extremely limited research budget, we felt that radical simplification of our experiments would allow us to demonstrate the feasibility of our ideas without requiring resources we simply did not have. Because many of the individual aspects that this project has brought together have been investigated with success, we were able to perform some experiments using proxies for the 'real thing'. | ||
| - | + | Please click [https://2010.igem.org/Team:NYU/Experiments Here] to see how we simulated each of the systems. | |
Latest revision as of 09:29, 27 October 2010

Our goal is to use the tools of synthetic biology to make a biological machine capable of antibody discovery. To do this, we have decided to learn from our own immune system by porting its own antibody discovery strategies into an engineered strain of yeast. This yeast will be capable of screening a library of antibodies against a target antigen, recombining the antibody gene in vivo and then either secreting or surface displaying the resulting antibody protein for a variety of purposes. Our hope is to demonstrate the feasibility of using the yeast cell not only as a vessel for antibody discovery but as a streamlined processing unit that can discover and also begin production of new antibodies - all in one test tube!
Project Details
Our plan for this year's competition is to construct an easy-to-use yeast strain capable of intracellular antibody discovery. The current dominant mode of microbial antibody discovery requires cellular surface display and high-throughput fluorescent screening. Instead of using this method, we wanted our cells to be able to sense if the antibody they are translating will bind the target antigen.
Using a modified form of the yeast two-hybrid screening system, the yeast cells are able to sense the amount of antibody::antigen interaction inside the cell. When the antibody binds the target antigen it anchors the VP16 transcriptional activator near our response system. VP16 then increases the level of transcription of a reporter gene. If this gene encodes a nutritional marker, such as Ura3, only cells experiencing antibody:antigen interactions can be selected using selective media. Using this method the original population of our yeast strain will be able to, in essence, screen itself and our resulting yeast population will contain only antibodies that bind our antigen.
In a research setting, once you have discovered the antibody you wish to use you must then reclone the gene for it into a secretion vector for production of pure antibody protein. This involves taking the plasmid out of the yeast cell, excising the antibody gene, ligating it into the second vector and then transforming it back into a suitable yeast strain.
To make this process faster and more easily accomplished, we have incorporated a recombination-based architecture into our system that will allow the cells to modify the antibody plasmid in vivo. Using this system transcription of the antibody gene can go straight from screening to protein production in a matter of minutes. This concept has the capability to not only shave weeks off of current antibody discovery protocols, but opens the door to cellular programming for other methods, such as restructuring of antibody genes or performing more complex screenings for different purposes.
The Experiments
In the spectrum of molecular biology, iGEM is a very short time frame. Because of this short time frame, our wet lab team having only two members and our extremely limited research budget, we felt that radical simplification of our experiments would allow us to demonstrate the feasibility of our ideas without requiring resources we simply did not have. Because many of the individual aspects that this project has brought together have been investigated with success, we were able to perform some experiments using proxies for the 'real thing'.
Please click Here to see how we simulated each of the systems.
 "
"