Team:NYU/Programming
From 2010.igem.org
(→Overlap Oligo Maker) |
|||
| (22 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<div id="allcontent"> | <div id="allcontent"> | ||
| - | <img class="bannerimage" src="https://static.igem.org/mediawiki/2010/ | + | <img class="bannerimage" src="https://static.igem.org/mediawiki/2010/c/ce/NYU_cavebanner_1.png" height="200px" width="950px"><br /> |
| Line 11: | Line 11: | ||
</li> | </li> | ||
| - | <li><a class="bannertoplinks" href=" | + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Project">Project</a> |
<ul style="z-index:1"> | <ul style="z-index:1"> | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Project">Overview</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Project">Overview</a></li> | ||
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Experiments">Experiments</a></li> | ||
| + | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Assembly">Overlap Assembly</a></li> | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Notebook">Notebook</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Notebook">Notebook</a></li> | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Parts">Biobricks</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Parts">Biobricks</a></li> | ||
| Line 19: | Line 21: | ||
</ul> | </ul> | ||
</li> | </li> | ||
| - | <li><a class="bannertoplinks" href=" | + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Team">Team</a> |
<ul style="z-index:1"> | <ul style="z-index:1"> | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Team">NYU</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Team">NYU</a></li> | ||
| Line 25: | Line 27: | ||
</ul> | </ul> | ||
</li> | </li> | ||
| - | <li><a class="bannertoplinks" href=" | + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a> |
<ul style="z-index:1"> | <ul style="z-index:1"> | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a></li> | ||
| - | <li><a class="bannerlinks" href=" | + | <li><a class="bannerlinks" href="http://www.sciencehouse.com">ScienceHouse</a></li> |
| + | <li><a class="bannerlinks" href="http://www.nysynbio.org">NY synbio</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| - | <li><a class="bannertoplinks" href=" | + | <li><a class="bannertoplinks" href="https://2010.igem.org/Team:NYU/Contact">Contact Us</a> |
| - | + | ||
| - | + | ||
| - | + | ||
</li> | </li> | ||
</ul> | </ul> | ||
| Line 174: | Line 175: | ||
padding: 3px 20px 3px 20px; | padding: 3px 20px 3px 20px; | ||
} | } | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
#sidebar { | #sidebar { | ||
| Line 282: | Line 254: | ||
</style> | </style> | ||
| - | <!-- !END NYU | + | <!-- !END NYU CSS- Thanks Harvard --> |
| + | |||
</html> | </html> | ||
| + | |||
| + | [[Image:NYU_OverviewSideF_2.png|200px|right]] | ||
=====About this Page===== | =====About this Page===== | ||
* Competent programming skills are quickly becoming a necessity for working in biology. Because most of our team members had never had exposure to a programming environment, we decided to use the obstacles presented in our iGEM experiments as cause for learning the Perl programming language and produced some simple scripts that could do routine iGEM labors for us. Here are some examples: | * Competent programming skills are quickly becoming a necessity for working in biology. Because most of our team members had never had exposure to a programming environment, we decided to use the obstacles presented in our iGEM experiments as cause for learning the Perl programming language and produced some simple scripts that could do routine iGEM labors for us. Here are some examples: | ||
| + | |||
=====Biobricking Primers===== | =====Biobricking Primers===== | ||
* This script allows the user to input a DNA sequence and outputs the primers necessary for biobricking that sequence via PCR. The script calculates Tm but does not (yet) consider primer dimers. | * This script allows the user to input a DNA sequence and outputs the primers necessary for biobricking that sequence via PCR. The script calculates Tm but does not (yet) consider primer dimers. | ||
| - | *Finally got the CGI working - go [http://russelldurrett.com/biobrickprimers.html | + | *Finally got the CGI working - click below or go [http://openwetware.org/images/a/a7/Biobrickprimers.txt here for the source code] |
| + | |||
| + | <html> | ||
| + | <a href="http://russelldurrett.com/biobrickprimers.html"><img src="https://static.igem.org/mediawiki/2010/7/77/Biobricking_Primers_Logo.gif"/></a> | ||
| + | </html> | ||
| + | |||
| - | |||
=====Overlap Oligo Maker===== | =====Overlap Oligo Maker===== | ||
| Line 300: | Line 280: | ||
* Some very fast and efficient DNA assembly methods require parts to overlap on the ends of their sequences. This script will take two DNA sequences and output the oligos (and their Tms) one would need to PCR constructs with overlapping ends. | * Some very fast and efficient DNA assembly methods require parts to overlap on the ends of their sequences. This script will take two DNA sequences and output the oligos (and their Tms) one would need to PCR constructs with overlapping ends. | ||
| - | *I'm a CGI machine! [http://www.russelldurrett.com/overlapoligos.html Check out the | + | *I'm a CGI machine! [http://openwetware.org/images/6/67/Overlapoligos.txt Here is the source code.] or click below for an interactive version. |
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="http://www.russelldurrett.com/overlapoligos.html"><img src="https://static.igem.org/mediawiki/2010/5/5c/Overlaplogo.gif"/></a></html> | ||
| + | |||
| + | |||
| + | =====Manipulation of Protein Structures===== | ||
| + | |||
| + | |||
| + | [[Image:Screen_shot_2010-10-27_at_10.19.59_PM.png|200px|left]] | ||
| + | |||
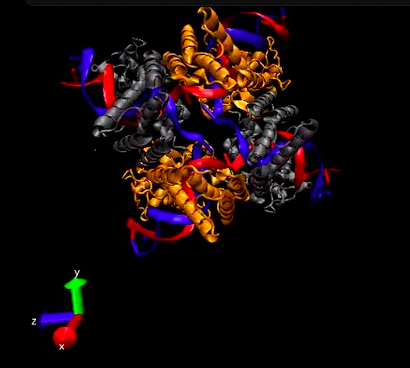
| + | In order to more fully understand certain parts of our system and to use the ridiculously amazing CAVE system at Cornell Med School, we decided to explore the 3D structures of the proteins involved in our system. Because the mechanism of Cre recombination is difficult to understand, we dove into the structures available on the Protein DataBase (PDB) and explored details of the mechanism. We were able to locate the active sites of the protein, isolate the catalytic residues, locate the vanadium cofactors and, in all, gain a better understanding of the protein's function. Check out some pictures below! | ||
| + | |||
| + | |||
| + | [[Image:Nyu_composite.png|400px|center]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =====Processing Visualization===== | ||
| + | |||
| + | |||
| + | Sung has also made a [http://www.processing.org Processing] language[[Image:Processing.jpg|200px|left]]-based visual aid describing the Immunoyeast system. You can go [http://www.russelldurrett.com/processing here] or click the icon below to interact with it to learn more about ImmunoYeast - don't forget to press all the buttons! | ||
| + | |||
| + | Also feel free to play around with the open source code! | ||
| + | |||
| + | |||
| + | We will also have an interactive touch-based system near our poster installation at the Jamboree, you won't want to miss it! | ||
| + | |||
| + | |||
| + | <html> | ||
| - | + | <a href="http://www.russelldurrett.com/processing" align="center"><img src="https://static.igem.org/mediawiki/2010/e/e5/Nyu_visualization.png"/></a></html> | |
Latest revision as of 03:59, 28 October 2010

Contents |
About this Page
- Competent programming skills are quickly becoming a necessity for working in biology. Because most of our team members had never had exposure to a programming environment, we decided to use the obstacles presented in our iGEM experiments as cause for learning the Perl programming language and produced some simple scripts that could do routine iGEM labors for us. Here are some examples:
Biobricking Primers
- This script allows the user to input a DNA sequence and outputs the primers necessary for biobricking that sequence via PCR. The script calculates Tm but does not (yet) consider primer dimers.
- Finally got the CGI working - click below or go here for the source code
Overlap Oligo Maker
- Some very fast and efficient DNA assembly methods require parts to overlap on the ends of their sequences. This script will take two DNA sequences and output the oligos (and their Tms) one would need to PCR constructs with overlapping ends.
- I'm a CGI machine! Here is the source code. or click below for an interactive version.
Manipulation of Protein Structures
In order to more fully understand certain parts of our system and to use the ridiculously amazing CAVE system at Cornell Med School, we decided to explore the 3D structures of the proteins involved in our system. Because the mechanism of Cre recombination is difficult to understand, we dove into the structures available on the Protein DataBase (PDB) and explored details of the mechanism. We were able to locate the active sites of the protein, isolate the catalytic residues, locate the vanadium cofactors and, in all, gain a better understanding of the protein's function. Check out some pictures below!
Processing Visualization
Sung has also made a Processing language-based visual aid describing the Immunoyeast system. You can go here or click the icon below to interact with it to learn more about ImmunoYeast - don't forget to press all the buttons!Also feel free to play around with the open source code!
We will also have an interactive touch-based system near our poster installation at the Jamboree, you won't want to miss it!
 "
"