Team:NYU/Experiments
From 2010.igem.org
(Difference between revisions)
| (One intermediate revision not shown) | |||
| Line 31: | Line 31: | ||
<li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a></li> | <li><a class="bannerlinks" href="https://2010.igem.org/Team:NYU/Sponsors">Sponsors</a></li> | ||
<li><a class="bannerlinks" href="http://www.sciencehouse.com">ScienceHouse</a></li> | <li><a class="bannerlinks" href="http://www.sciencehouse.com">ScienceHouse</a></li> | ||
| - | <li><a class="bannerlinks" href="http://nysynbio.org">NY synbio</a></li> | + | <li><a class="bannerlinks" href="http://www.nysynbio.org">NY synbio</a></li> |
</ul> | </ul> | ||
</li> | </li> | ||
| Line 259: | Line 259: | ||
| + | =====Antibody Screening===== | ||
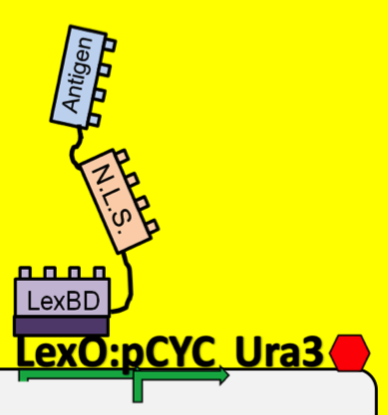
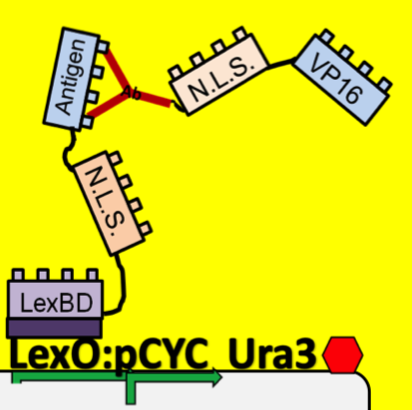
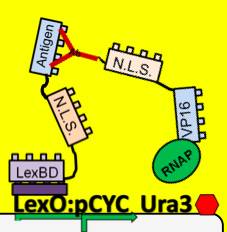
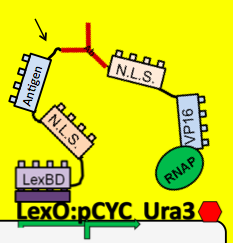
| - | + | *To see which antibodies in our diverse population are binding the antigen, we put our population through a modified yeast two-hybrid screen. As you see below, the LexBD domain binds lexO just upstream of the Ura3 CDS, anchoring the bait antigen to the DNA at that point. If the antibody binds the antigen, it localizes the fused VP16 domain to that site as well. This causes an increase of transcription of Ura3 which allows the cells with positive binding antibodies to survive in media without Uracil while those with antibodies that do not bind the antigen die off and are cleared from the population. | |
| - | + | ||
| - | + | ||
| - | + | ||
[[image:NYU_bind1.png|300px|left]] | [[image:NYU_bind1.png|300px|left]] | ||
| Line 270: | Line 268: | ||
[[image:NYU_bind3.png|300px|left]] | [[image:NYU_bind3.png|300px|left]] | ||
| - | |||
| - | |||
| - | [[image: | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | *Because iGEM is such a [[Image:NYU_binding4.png|300px|right]] short competition and biology takes time to engineer we decided to simulate this two-hybrid screen and ensure that a uracil selection system such as this would actually work. To do this, we physically joined the antibody and antigen constructs together by replacing those parts with a flexible (Gly3Ser)4 linker and nuclear localization sequences. The translated protein then becomes LexBD:NLS:linker:NLS:VP16. Because VP16 is now physically linked to the Lex DNA binding domain, as long as this construct is localized in the nucleus then VP16 will be anchored to the lex operator, increasing transcription through any promoter system that is in lexO's proximity. Because the screening no longer relies on antibody:antigen interactions, expression of this construct can serve as a positive control for the screen. We linked this construct to the pGAL promoter so it would only be expressed in galactose-containing media. To actually test if the system is working or not, we plate these cells on multiple -ura plates, some containing galactose and others containing only glucose. If the screening functions properly, the cells on the galactose plate should grow while the cells on the glucose plate die. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =====CRE Recombination===== | ||
| + | |||
| + | *To test for functionality of our Cre recombination constructs we built a reporter plasmid that can function as a positive control. [[image:NYU_plasmid.png|300px|left]] Because our system is all about the protein localization, secretion and surface display of proteins of interest we decided that our control plasmid should involve the same parts. We built a plasmid nearly identical to the [[image:NYU_creillustr.png|300px|right]] antibody plasmid in our system but substituted a yeast-optimized eCFP biobrick for the antibody gene. | ||
| + | |||
| + | At the outset of this system we observe the eCFP protein localized into the nucleus. Upon addition of galactose Cre recombinase linked to a nuclear localization sequence should be transcribed off of the Gal promoter, translated in the cytoplasm and then imported into the nucleus of the cell. There it recombines the plasmid between the two loxP sites. This brings the eCFP into a fusion with the secretion tag rather than the nuclear localization tag, so we should observe an eventual exclusion of CFP from the nucleus and secretion from the cell. | ||
Latest revision as of 03:37, 28 October 2010

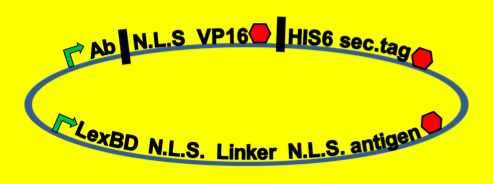
Antibody Screening
- To see which antibodies in our diverse population are binding the antigen, we put our population through a modified yeast two-hybrid screen. As you see below, the LexBD domain binds lexO just upstream of the Ura3 CDS, anchoring the bait antigen to the DNA at that point. If the antibody binds the antigen, it localizes the fused VP16 domain to that site as well. This causes an increase of transcription of Ura3 which allows the cells with positive binding antibodies to survive in media without Uracil while those with antibodies that do not bind the antigen die off and are cleared from the population.
- Because iGEM is such a short competition and biology takes time to engineer we decided to simulate this two-hybrid screen and ensure that a uracil selection system such as this would actually work. To do this, we physically joined the antibody and antigen constructs together by replacing those parts with a flexible (Gly3Ser)4 linker and nuclear localization sequences. The translated protein then becomes LexBD:NLS:linker:NLS:VP16. Because VP16 is now physically linked to the Lex DNA binding domain, as long as this construct is localized in the nucleus then VP16 will be anchored to the lex operator, increasing transcription through any promoter system that is in lexO's proximity. Because the screening no longer relies on antibody:antigen interactions, expression of this construct can serve as a positive control for the screen. We linked this construct to the pGAL promoter so it would only be expressed in galactose-containing media. To actually test if the system is working or not, we plate these cells on multiple -ura plates, some containing galactose and others containing only glucose. If the screening functions properly, the cells on the galactose plate should grow while the cells on the glucose plate die.
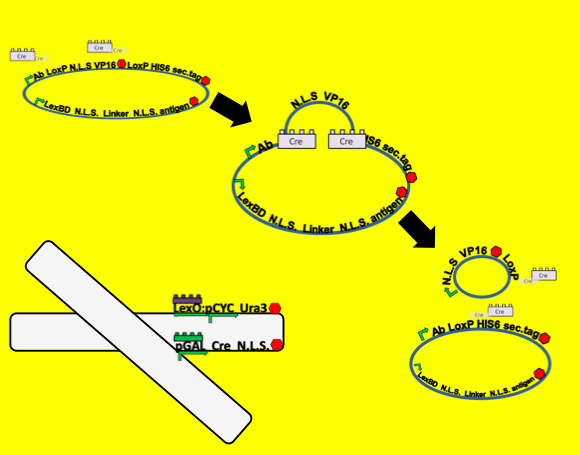
CRE Recombination
- To test for functionality of our Cre recombination constructs we built a reporter plasmid that can function as a positive control. Because our system is all about the protein localization, secretion and surface display of proteins of interest we decided that our control plasmid should involve the same parts. We built a plasmid nearly identical to the antibody plasmid in our system but substituted a yeast-optimized eCFP biobrick for the antibody gene.
At the outset of this system we observe the eCFP protein localized into the nucleus. Upon addition of galactose Cre recombinase linked to a nuclear localization sequence should be transcribed off of the Gal promoter, translated in the cytoplasm and then imported into the nucleus of the cell. There it recombines the plasmid between the two loxP sites. This brings the eCFP into a fusion with the secretion tag rather than the nuclear localization tag, so we should observe an eventual exclusion of CFP from the nucleus and secretion from the cell.
 "
"