From 2010.igem.org
(Difference between revisions)
Revision as of 17:13, 26 October 2010
Fusion Protein
We need to fusion protein blah blah blah
Legend
| Effect | Class
|
| Primers: (changes overline/underline and background)
|
| Forward Primer | FP
|
| Overlapping Primer | OP
|
| Reverse Primer | RP
|
| Parts: (changes text color)
|
| Promoter | P
|
| Ribosome Binding site | R
|
| RNA | rna
|
| Coding region | C
|
| Terminator | T
|
| Special | S
|
| Cutting sites: (changes background)
|
| Biobrick cutting site | cut
|
| Biobrick cutting site mutation | cutmut
|
GFP
We based splitting GFP ([http://partsregistry.org/Part:BBa_E0040 BBa_E0040]) based on the split point at 157&158aa used by iGEM07_Davidson_Missouri's [http://partsregistry.org/Part:BBa_I715019 BBa_I715019] and [http://partsregistry.org/Part:BBa_I715020 BBa_I715020]. The A-part split is the same as [http://partsregistry.org/Part:BBa_I715019 BBa_I715019], but the B-part has one base difference from [http://partsregistry.org/Part:BBa_I715020 BBa_I715020].
| >GFP ([http://partsregistry.org/Part:BBa_E0040 BBa_E0040]) | 720bp |
atgcgtaaaggagaagaacttttcactggagttgtcccaattcttgttgaattagatggtgatgttaatgggcacaaattttctgtcagtggaga
gggtgaaggtgatgcaacatacggaaaacttacccttaaatttatttgcactactggaaaactacctgttccatggccaacacttgtcactactt
tcggttatggtgttcaatgctttgcgagatacccagatcatatgaaacagcatgactttttcaagagtgccatgcccgaaggttatgtacaggaa
agaactatatttttcaaagatgacgggaactacaagacacgtgctgaagtcaagtttgaaggtgatacccttgttaatagaatcgagttaaaagg
tattgattttaaagaagatggaaacattcttggacacaaattggaatacaactataactcacacaatgtatacatcatggcagacaaacaaaaga
atggaatcaaagttaacttcaaaattagacacaacattgaagatggaagcgttcaactagcagaccattatcaacaaaatactccaattggcgat
ggccctgtccttttaccagacaaccattacctgtccacacaatctgccctttcgaaagatcccaacgaaaagagagaccacatggtccttcttga
gtttgtaacagctgctgggattacacatggcatggatgaactatacaaataataa
| >GFP_A ([http://partsregistry.org/Part:BBa_I175019 BBa_I175019]) | 471bp |
atgcgtaaaggagaagaacttttcactggagttgtcccaattcttgttgaattagatggtgatgttaatgggcacaaattttctgtcagtggaga
gggtgaaggtgatgcaacatacggaaaacttacccttaaatttatttgcactactggaaaactacctgttccatggccaacacttgtcactactt
tcggttatggtgttcaatgctttgcgagatacccagatcatatgaaacagcatgactttttcaagagtgccatgcccgaaggttatgtacaggaa
agaactatatttttcaaagatgacgggaactacaagacacgtgctgaagtcaagtttgaaggtgatacccttgttaatagaatcgagttaaaagg
tattgattttaaagaagatggaaacattcttggacacaaattggaatacaactataactcacacaatgtatacatcatggcagacaaacaa
FP (including start codon): atgcgtaaaggagaagaacttttc (55c,24bp,38gc)
RP: ttgtttgtctgccatgatgtatac (55c,24bp,38gc)
aagaatggaatcaaagttaacttcaaaattagacacaacattgaagatggaagcgttcaactagcagaccattatcaacaaaatactccaattg
gcgatggccctgtccttttaccagacaaccattacctgtccacacaatctgccctttcgaaagatcccaacgaaaagagagaccacatggtcctt
cttgagtttgtaacagctgctgggattacacatggcatggatgaactatacaaataataa
FP: aagaatggaatcaaagttaacttcaaaa (55c,28bp,25gc)
RP (excluding stop codon): tttgtatagttcatccatgccatg (55c,24bp,38gc)
RFP
We based splitting RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) based on the split point at 154&155aa used by iGEM07_Davidson_Missouri's [http://partsregistry.org/Part:BBa_I715022 BBa_I715022] and [http://partsregistry.org/Part:BBa_I715023 BBa_I715023]. The A-part split is the same as [http://partsregistry.org/Part:BBa_I715022 BBa_I715022], but the B-part has one base difference from [http://partsregistry.org/Part:BBa_I715023 BBa_I715023].
| >RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) | 681bp |
atggcttcctccgaagacgttatcaaagagttcatgcgtttcaaagttcgtatggaaggttccgttaacggtcacgagttcgaaatcgaaggtgaaggtg
aaggtcgtccgtacgaaggtacccagaccgctaaactgaaagttaccaaaggtggtccgctgccgttcgcttgggacatcctgtccccgcagttccagta
cggttccaaagcttacgttaaacacccggctgacatcccggactacctgaaactgtccttcccggaaggtttcaaatgggaacgtgttatgaacttcgaa
gacggtggtgttgttaccgttacccaggactcctccctgcaagacggtgagttcatctacaaagttaaactgcgtggtaccaacttcccgtccgacggtc
cggttatgcagaaaaaaaccatgggttgggaagcttccaccgaacgtatgtacccggaagacggtgctctgaaaggtgaaatcaaaatgcgtctgaaact
gaaagacggtggtcactacgacgctgaagttaaaaccacctacatggctaaaaaaccggttcagctgccgggtgcttacaaaaccgacatcaaactggac
atcacctcccacaacgaagactacaccatcgttgaacagtacgaacgtgctgaaggtcgtcactccaccggtgcttaataa
| >RFP_A ([http://partsregistry.org/Part:BBa_I715022 BBa_I715022]) | 462bp |
atggcttcctccgaagacgttatcaaagagttcatgcgtttcaaagttcgtatggaaggttccgttaacggtcacgagttcgaaatcgaaggtgaaggtg
aaggtcgtccgtacgaaggtacccagaccgctaaactgaaagttaccaaaggtggtccgctgccgttcgcttgggacatcctgtccccgcagttccagta
cggttccaaagcttacgttaaacacccggctgacatcccggactacctgaaactgtccttcccggaaggtttcaaatgggaacgtgttatgaacttcgaa
gacggtggtgttgttaccgttacccaggactcctccctgcaagacggtgagttcatctacaaagttaaactgcgtggtaccaacttcccgtccgacggtc
cggttatgcagaaaaaaaccatgggttgggaagcttccaccgaacgtatgtacccggaagac
FP (including start codon): atggcttcctccgaagac (55C,18bp,56%)
RP: gtcttccgggtacatacgtt (55C,20bp,50%)
ggtgctctgaaaggtgaaatcaaaatgcgtctgaaactgaaagacggtggtcactacgacgctgaagttaaaaccacctacatggctaaaaaaccggttc
agctgccgggtgcttacaaaaccgacatcaaactggacatcacctcccacaacgaagactacaccatcgttgaacagtacgaacgtgctgaaggtcgtca
ctccaccggtgcttaataa
FP: ggtgctctgaaaggtgaaatc (55C,21bp,48%)
RP (excluding stop codon): agcaccggtggagtga (56C,16bp,63%)
eIF4A
We split eIF4A ([http://partsregistry.org/Part:BBa_K411100 BBa_K411100]) at 215&216aa based on [http://igem.ym.edu.tw/pv2010/images/b/be/File:NYMU_Nmeth1023-S5.pdf 2.protocol file from Natalia E. Broude]. But first we needed to mutate the two PstI cutting sites.
We took the protein coding region from the [http://www.ncbi.nlm.nih.gov/nuccore/NM_144958 eIF4A mRNA transcript sequence from Mouse (from NCBI)] and found it had 2 PstI cutting sites:
| >eIF4A_original([http://partsregistry.org/Part:BBa_K411100 BBa_K411100]) | 1173bp |
atggagccggaaggcgtcatcgagagtaactggaacgagattgtggatagctttgatgacatgaatctctcagagtccctcctccgtggtatttatgcct
atggttttgagaagccctctgccatccagcagcgagctattcttccttgtatcaagggttatgatgtgattgctcaagcccagtctgggactgggaaaac
agctacatttgccatatcaattctgcagcagattgaattagatctaaaggccactcaggctttggttctggcacccacacgtgaattggctcagcagata
caaaaggtggttatggcattaggagactacatgggtgcctcttgtcatgcctgcattgggggcaccaatgtgcgtgctgaggtgcagaagctgcagatgg
aagctccccatatcatcgtgggtacccctggccgggtgtttgacatgcttaaccggagatacctgtcccccaaatacatcaagatgttcgtactggatga
agcagatgaaatgttaagccgagggttcaaggatcagatctatgacatattccagaagctcaacagcaacacacaggtagttttgttgtctgctacaatg
ccttctgatgtccttgaggtgaccaagaaatttatgagagaccctattcggattcttgtcaagaaggaagaattgaccctggagggtatccgccaattct
acatcaatgtggaacgagaggagtggaagcttgacacattgtgtgacttgtatgagacgctgaccatcacccaggcagtcatctttatcaacaccagaag
gaaggtggactggctcaccgagaagatgcatgcccgagatttcactgtttctgccatgcacggagatatggaccaaaaggaacgagatgtgatcatgagg
gagttccggtctggctctagcagagtattaattaccactgacctgttggccagaggcattgatgtgcagcaggtctccttagtcatcaactatgaccttc
ccaccaacagggaaaactacatccacagaatcggtcgaggtggtcggtttggtcgtaagggtgtggctattaacatggtgaccgaagaagacaagaggac
tcttcgagacattgagactttctacaacacctccattgaagagatgcccctcaacgttgctgacctcatttga
We need to mutate out the two PstI cutting sites. The template of eIF4A on a [http://genome-www.stanford.edu/vectordb/vector_descrip/COMPLETE/PGEX4T1.SEQ.html pGEX-4TI vector] was kindly provided by C.Proud. We used these primers to mutate the cutting site:
2 of pstI cutting site
mut1(24bp,55C,42%)
FP: ccatatcaattctccagcagattga
RP: caatctgctggagaattgatatggc
mut2(18bp,55C,56%)
FP: gcagaagctccagatggaa
RP: tccatctggagcttctgca
The new sequence after mutation:
| >eIF4A ([http://partsregistry.org/Part:BBa_K411100 BBa_K411100]) | 1173bp |
atggagccggaaggcgtcatcgagagtaactggaacgagattgtggatagctttgatgacatgaatctctcagagtccctcctccgtggtatttatgcct
atggttttgagaagccctctgccatccagcagcgagctattcttccttgtatcaagggttatgatgtgattgctcaagcccagtctgggactgggaaaac
agctacatttgccatatcaattctccagcagattgaattagatctaaaggccactcaggctttggttctggcacccacacgtgaattggctcagcagata
caaaaggtggttatggcattaggagactacatgggtgcctcttgtcatgcctgcattgggggcaccaatgtgcgtgctgaggtgcagaagctccagatgg
aagctccccatatcatcgtgggtacccctggccgggtgtttgacatgcttaaccggagatacctgtcccccaaatacatcaagatgttcgtactggatga
agcagatgaaatgttaagccgagggttcaaggatcagatctatgacatattccagaagctcaacagcaacacacaggtagttttgttgtctgctacaatg
ccttctgatgtccttgaggtgaccaagaaatttatgagagaccctattcggattcttgtcaagaaggaagaattgaccctggagggtatccgccaattct
acatcaatgtggaacgagaggagtggaagcttgacacattgtgtgacttgtatgagacgctgaccatcacccaggcagtcatctttatcaacaccagaag
gaaggtggactggctcaccgagaagatgcatgcccgagatttcactgtttctgccatgcacggagatatggaccaaaaggaacgagatgtgatcatgagg
gagttccggtctggctctagcagagtattaattaccactgacctgttggccagaggcattgatgtgcagcaggtctccttagtcatcaactatgaccttc
ccaccaacagggaaaactacatccacagaatcggtcgaggtggtcggtttggtcgtaagggtgtggctattaacatggtgaccgaagaagacaagaggac
tcttcgagacattgagactttctacaacacctccattgaagagatgcccctcaacgttgctgacctcatttga
atggagccggaaggcgtcatcgagagtaactggaacgagattgtggatagctttgatgacatgaatctctcagagtccctcctccgtggtatttatgcct
atggttttgagaagccctctgccatccagcagcgagctattcttccttgtatcaagggttatgatgtgattgctcaagcccagtctgggactgggaaaac
agctacatttgccatatcaattctccagcagattgaattagatctaaaggccactcaggctttggttctggcacccacacgtgaattggctcagcagata
caaaaggtggttatggcattaggagactacatgggtgcctcttgtcatgcctgcattgggggcaccaatgtgcgtgctgaggtgcagaagctccagatgg
aagctccccatatcatcgtgggtacccctggccgggtgtttgacatgcttaaccggagatacctgtcccccaaatacatcaagatgttcgtactggatga
agcagatgaaatgttaagccgagggttcaaggatcagatctatgacatattccagaagctcaacagcaacacacaggtagttttgttgtctgctacaatg
ccttctgatgtccttgaggtgaccaagaaatttatgagagaccct
FP: atggagccggaaggcgtcatcga (66c,24bp,61gc) [design error -- was suppose to design it to be 55c]
RP: agggtctctcataaatttcttgg (54C,23bp,39%)
attcggattcttgtcaagaaggaagaattgaccctggagggtatccgccaattct
acatcaatgtggaacgagaggagtggaagcttgacacattgtgtgacttgtatgagacgctgaccatcacccaggcagtcatctttatcaacaccagaag
gaaggtggactggctcaccgagaagatgcatgcccgagatttcactgtttctgccatgcacggagatatggaccaaaaggaacgagatgtgatcatgagg
gagttccggtctggctctagcagagtattaattaccactgacctgttggccagaggcattgatgtgcagcaggtctccttagtcatcaactatgaccttc
ccaccaacagggaaaactacatccacagaatcggtcgaggtggtcggtttggtcgtaagggtgtggctattaacatggtgaccgaagaagacaagaggac
tcttcgagacattgagactttctacaacacctccattgaagagatgcccctcaacgttgctgacctcatttga
FP: attcggattcttgtcaagaagg (54C,22bp,41%)
RP (including stop codon): tcaaatgaggtcagcaacgttgag (59c,24bp,46gc) [design error -- was suppose to design it to be 55c]
Linker
We took the linker from 2.protocol file from Natalia E. Broude:[http://igem.ym.edu.tw/pv2010/images/b/be/File:NYMU_Nmeth1023-S5.pdf ]
ggcagcagcggcagcagcggcagcggcagc
We decided to use primers to create the linker:
cggcagcagcggcagcggcagc
ggcagcagcggcagcagcggcagcggcagc
ggcagcagcggcagcagcggcagc
overlapping region: cggcagcagcggcagc (63c,16bp,81gc)
FP: cggcagcagcggcagcggcagc (22bp)
RP: gctgccgctgctgccgctgctgcc (24bp)
Fusioning it together
GFP fusion part
(note the extra taa stop codon at the end)
| >GFP-linker-eIF4A_A ([http://partsregistry.org/Part:BBa_K411101 BBa_K411101]) | 1149bp |
atgcgtaaaggagaagaacttttcactggagttgtcccaattcttgttgaattagatggtgatgttaatgggcacaaattttctgtcagtggagagggtg
aaggtgatgcaacatacggaaaacttacccttaaatttatttgcactactggaaaactacctgttccatggccaacacttgtcactactttcggttatgg
tgttcaatgctttgcgagatacccagatcatatgaaacagcatgactttttcaagagtgccatgcccgaaggttatgtacaggaaagaactatatttttc
aaagatgacgggaactacaagacacgtgctgaagtcaagtttgaaggtgatacccttgttaatagaatcgagttaaaaggtattgattttaaagaagatg
gaaacattcttggacacaaattggaatacaactataactcacacaatgtatacatcatggcagacaaacaaggcagcagcggcagcagcggcagcggcag
catggagccggaaggcgtcatcgagagtaactggaacgagattgtggatagctttgatgacatgaatctctcagagtccctcctccgtggtatttatgcc
tatggttttgagaagccctctgccatccagcagcgagctattcttccttgtatcaagggttatgatgtgattgctcaagcccagtctgggactgggaaaa
cagctacatttgccatatcaattctccagcagattgaattagatctaaaggccactcaggctttggttctggcacccacacgtgaattggctcagcagat
acaaaaggtggttatggcattaggagactacatgggtgcctcttgtcatgcctgcattgggggcaccaatgtgcgtgctgaggtgcagaagctccagatg
gaagctccccatatcatcgtgggtacccctggccgggtgtttgacatgcttaaccggagatacctgtcccccaaatacatcaagatgttcgtactggatg
aagcagatgaaatgttaagccgagggttcaaggatcagatctatgacatattccagaagctcaacagcaacacacaggtagttttgttgtctgctacaat
gccttctgatgtccttgaggtgaccaagaaatttatgagagacccttga
GFP_split_A_FP: gaattcgcggccgcttctagag atgcgtaaaggagaagaacttttc (46bp) OK
GFP_split_A_RP: gctgccgctgctgccgctgctgcc ttgtttgtctgccatgatgtatac (48bp) RE DING
eIF4A_split_A_FP: cggcagcagcggcagcggcagc atggagccggaaggcgtcatcga (45bp) OK
eIF4A_split_A_RP: ctgcagcggccgctactagta tca agggtctctcataaatttcttgg (47bp) OK
(note that we added as start codon and removed the stop codon of GFP_B)
| >GFP-linker-eIF4A_B ([http://partsregistry.org/Part:BBa_K411102 BBa_K411102]) | 804bp |
atgaagaatggaatcaaagttaacttcaaaattagacacaacattgaagatggaagcgttcaactagcagaccattatcaacaaaatactccaattggcg
atggccctgtccttttaccagacaaccattacctgtccacacaatctgccctttcgaaagatcccaacgaaaagagagaccacatggtccttcttgagtt
tgtaacagctgctgggattacacatggcatggatgaactatacaaaggcagcagcggcagcagcggcagcggcagcattcggattcttgtcaagaaggaa
gaattgaccctggagggtatccgccaattctacatcaatgtggaacgagaggagtggaagcttgacacattgtgtgacttgtatgagacgctgaccatca
cccaggcagtcatctttatcaacaccagaaggaaggtggactggctcaccgagaagatgcatgcccgagatttcactgtttctgccatgcacggagatat
ggaccaaaaggaacgagatgtgatcatgagggagttccggtctggctctagcagagtattaattaccactgacctgttggccagaggcattgatgtgcag
caggtctccttagtcatcaactatgaccttcccaccaacagggaaaactacatccacagaatcggtcgaggtggtcggtttggtcgtaagggtgtggcta
ttaacatggtgaccgaagaagacaagaggactcttcgagacattgagactttctacaacacctccattgaagagatgcccctcaacgttgctgacctcat
ttga
GFP_split_B_FP: gaattcgcggccgcttctagag atg aagaatggaatcaaagttaacttcaaaa (53bp)
GFP_split_B_RP: gctgccgctgctgccgctgctgcc tttgtatagttcatccatgccatg (48bp)
eIF4A_split_B_FP: cggcagcagcggcagcggcagc attcggattcttgtcaagaagg (44bp)
eIF4A_split_B_RP: ctgcagcggccgctactagta tcaaatgaggtcagcaacgttgag (45bp)
RFP fusion part
(note the extra taa stop codon at the end)
| >RFP-linker-eIF4A_A ([http://partsregistry.org/Part:BBa_K411103 BBa_K411103]) | 1140bp |
atggcttcctccgaagacgttatcaaagagttcatgcgtttcaaagttcgtatggaaggttccgttaacggtcacgagttcgaaatcgaaggtgaaggtg
aaggtcgtccgtacgaaggtacccagaccgctaaactgaaagttaccaaaggtggtccgctgccgttcgcttgggacatcctgtccccgcagttccagta
cggttccaaagcttacgttaaacacccggctgacatcccggactacctgaaactgtccttcccggaaggtttcaaatgggaacgtgttatgaacttcgaa
gacggtggtgttgttaccgttacccaggactcctccctgcaagacggtgagttcatctacaaagttaaactgcgtggtaccaacttcccgtccgacggtc
cggttatgcagaaaaaaaccatgggttgggaagcttccaccgaacgtatgtacccggaagacggcagcagcggcagcagcggcagcggcagcatggagcc
ggaaggcgtcatcgagagtaactggaacgagattgtggatagctttgatgacatgaatctctcagagtccctcctccgtggtatttatgcctatggtttt
gagaagccctctgccatccagcagcgagctattcttccttgtatcaagggttatgatgtgattgctcaagcccagtctgggactgggaaaacagctacat
ttgccatatcaattctccagcagattgaattagatctaaaggccactcaggctttggttctggcacccacacgtgaattggctcagcagatacaaaaggt
ggttatggcattaggagactacatgggtgcctcttgtcatgcctgcattgggggcaccaatgtgcgtgctgaggtgcagaagctccagatggaagctccc
catatcatcgtgggtacccctggccgggtgtttgacatgcttaaccggagatacctgtcccccaaatacatcaagatgttcgtactggatgaagcagatg
aaatgttaagccgagggttcaaggatcagatctatgacatattccagaagctcaacagcaacacacaggtagttttgttgtctgctacaatgccttctga
tgtccttgaggtgaccaagaaatttatgagagacccttga
RFP_split_A_FP: gaattcgcggccgcttctag atggcttcctccgaagac (38bp)
RFP_split_A_RP: gctgccgctgctgccgctgctgcc gtcttccgggtacatacgtt (44bp)
eIF4A_split_A_FP and RP are the same as the GFP part above.
(note that we added as start codon and removed the stop codon of RFP_B)
| >RFP-linker-eIF4A_B ([http://partsregistry.org/Part:BBa_K411104 BBa_K411104]) | 774bp |
atgggtgctctgaaaggtgaaatcaaaatgcgtctgaaactgaaagacggtggtcactacgacgctgaagttaaaaccacctacatggctaaaaaaccgg
ttcagctgccgggtgcttacaaaaccgacatcaaactggacatcacctcccacaacgaagactacaccatcgttgaacagtacgaacgtgctgaaggtcg
tcactccaccggtgctggcagcagcggcagcagcggcagcggcagcattcggattcttgtcaagaaggaagaattgaccctggagggtatccgccaattc
tacatcaatgtggaacgagaggagtggaagcttgacacattgtgtgacttgtatgagacgctgaccatcacccaggcagtcatctttatcaacaccagaa
ggaaggtggactggctcaccgagaagatgcatgcccgagatttcactgtttctgccatgcacggagatatggaccaaaaggaacgagatgtgatcatgag
ggagttccggtctggctctagcagagtattaattaccactgacctgttggccagaggcattgatgtgcagcaggtctccttagtcatcaactatgacctt
cccaccaacagggaaaactacatccacagaatcggtcgaggtggtcggtttggtcgtaagggtgtggctattaacatggtgaccgaagaagacaagagga
ctcttcgagacattgagactttctacaacacctccattgaagagatgcccctcaacgttgctgacctcatttga
RFP_split_B_FP: gaattcgcggccgcttctag atg ggtgctctgaaaggtgaaatc (44bp)
RFP_split_B_RP: gctgccgctgctgccgctgctgcc agcaccggtggagtga (40bp)
eIF4A_split_B_FP and RP are the same as the GFP part above.
Aptamer
- Aptamer in front of non-coding sequence:
-
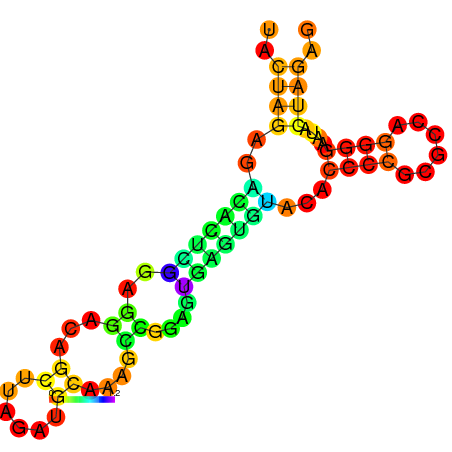
RNAfold prediction of Aptamer Structure with Biobrick scars in front of a non-coding region. The scars bond together, but the overall structure does not change.
- Check to see if the addition of the biobrick scars affect the secondary structure in front of coding sequence:
-
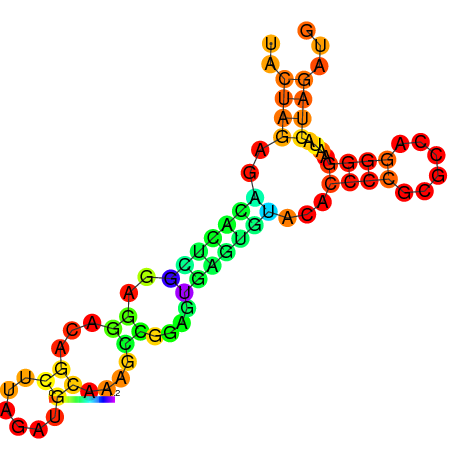
RNAfold prediction of Aptamer Structure with Biobrick scars in front of a coding region. The scars bond together, but the overall structure does not change.
- Structure interacting with the split proteins:
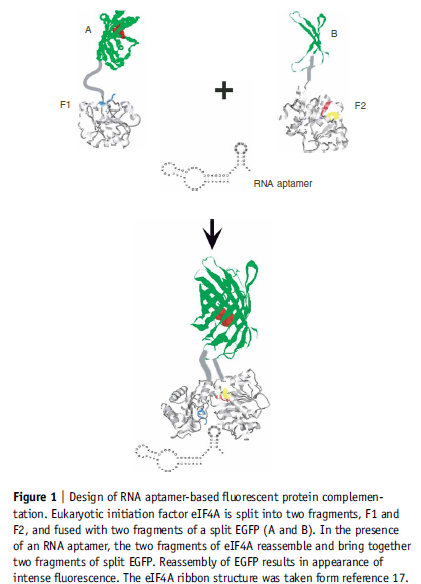
Aptamer Structure interacting with split protein [6]
 "
"


