Team:Michigan/Virus Surface Display
From 2010.igem.org
|
7/10/2010 13:00 • digest I719015 (T7 GFPmut3b) and I719005(T7 Promoter) with EcoR1 and Pst1.
• inoculate competent cell culture • harvest I719015, K145001, K117008, K117002, K103006 from registry
• streak plate INP NC sample
• miniprep INP NC sample
• digest INP NC with EcoRI and SpeI
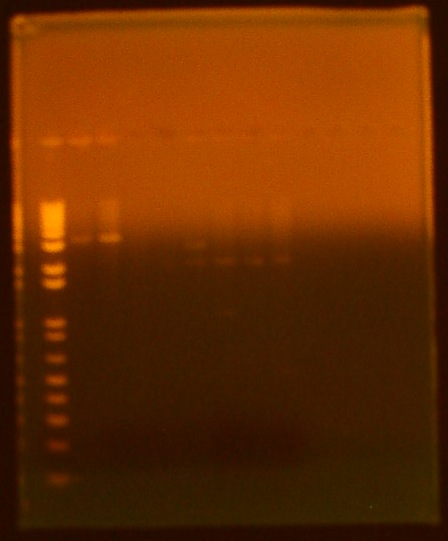
• gel electrophoresis of all biobricks lane 1 - ladder lane 2 - INP NC sample 1 (K265008) lane 3 - INP NC sample 2 (K265008) lane 4 - T7 GFPmut3b (I719015) lane 5 - T7 Promoter (I719005) lane 6 - T7 RNA polymerase (K145001) lane 7 - pLsrA-YFP (K117008) lane 8 - LsrA promoter (K117002) lane 9 - OmpA (K103006)
• aliquot INP samples and primers for sequencing
• Electroporation transformation of the following Biobrick parts: 1. pBad/araC (I0500) 2. Linker (K157013) 3. GFP (E0040)
• miniprep
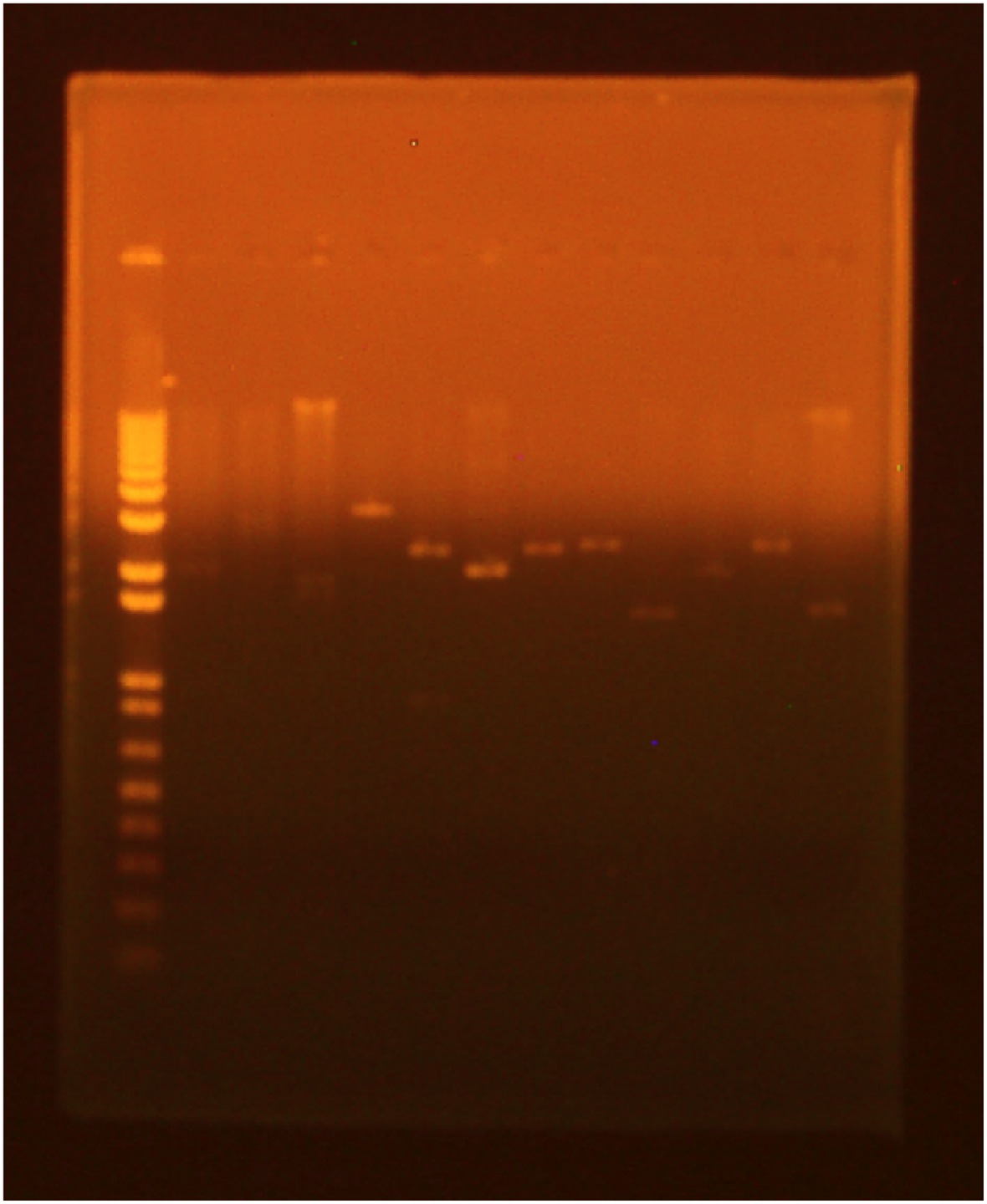
• digest • gel electrophoresis Lane 1 (far right): Invitrogen 1 kb Plus ladder Lane 2: GFP cut with XbaI and PstI Lane 3: GFP cut with EcoRI and XbaI Lane 4: uncut GFP plamid Lane 5:INP cut with EcoRI and SpeI Lane 6:INP cut with SpeI and PstI Lane 7:uncut INP plasmid Lane 8:Linker cut with XbaI and SpeI Lane 9:Linker cut with SpeI and PstI Lane 10: uncut Linker plasmid Lane 11: OmpA cut with EcoRI and SpeI Lane 12: OmpA cut with SpeI and PstI Lane 13:uncut OmpA plasmid
• Ligation of digested Biobrick parts: - INP + Linker - OmpA + GFP - INP + GFP • Heat shock transformation of ligation
INP + Linker construct ng/uL OD260/280 OD260/230
1.1 20.7 1.88 1.73
1.2 61.4 2.04 2.09
2.1 59.8 1.99 2.06
2.2 69.8 2.02 2.11
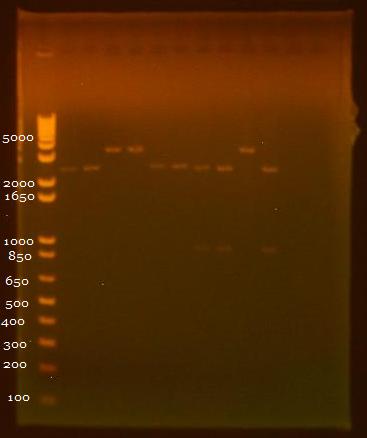
8/3/2010 • EcoRI+SpeI and SpeI+PstI digests of INP + linker minipreps • Gel electrophoresis results: Lane 1 - 1 kb Plus Ladder Lane 2 - INP + Linker 1.1 (EcoRI+SpeI) Lane 3 - INP + Linker 1.2 (EcoRI+SpeI) Lane 4 - INP + Linker 2.1 (EcoRI+SpeI) Lane 5 - INP + Linker 2.2 (EcoRI+SpeI) Lane 6 - INP + Linker 1.1 (SpeI+PstI) Lane 7 - INP + Linker 1.2 (SpeI+PstI) Lane 8 - INP + Linker 2.1 (SpeI+PstI) Lane 9 - INP + Linker 2.2 (SpeI+PstI) Lane 10 - INP 1 (EcoRI+SpeI) Lane 11 - INP 2 (SpeI+PstI)
• Inoculated 6 ml DH5α for ligation and transformation.
• OmpA and GFP Ligations done: 1. OmpA1 (insert) + GFP2 (backbone) 2. OmpA2 (backbone) + GFP1 (insert) • Heat shock transformation
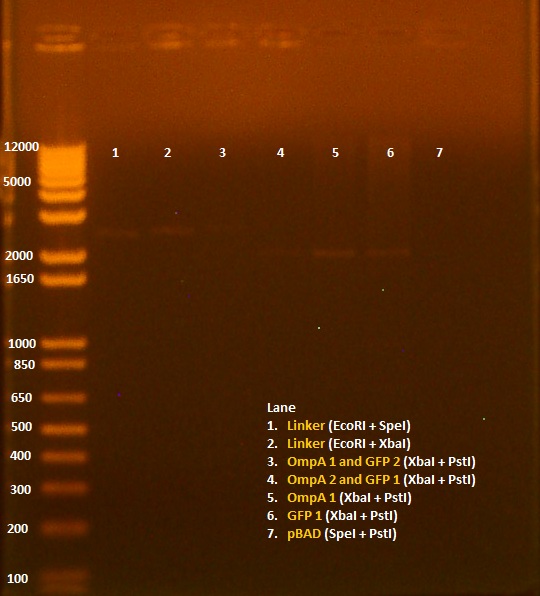
• Miniprep of 8/6 transformation, in addition to pBAD promoter and linker. • Nanodrop results: Sample OD260/280 OD260/230 ng/uL 1. pBAD 1.72 8.91 11.5 2. OmpA+GFP 1 2.88 -3.52 4.5 3. OmpA+GFP 2 1.62 3.03 7.9 4. Linker 2.10 -12.00 9.4 • 20 uL digest without water - 5 uL NEB2 buffer - 0.5 uL 100X BSA - 1 uL of each enzyme - 12.5 uL of DNA • Digests: 1. Linker 1 (EcoRI+SpeI) 2. Linker 2 (EcoRI+SpeI) 3. OmpA+GFP 1 (XbaI+PstI) 4. OmpA+GFP 2 (XbaI+PstI) 5. pBAD (SpeI+PstI)
• Gel of 8/9 digest results
• Due to poor digest results, we repeated the miniprep from 8/9, with additional colonies of OmpA+GFP picked. Also miniprepped are 2 expression plasmids (Backbone and Promoter) transformed earlier by another group. Sample
|
In the Lab
|
 "
"