Team:Mexico-UNAM-CINVESTAV/Notebook/Week Four
From 2010.igem.org
(→Wenesday) |
|||
| (2 intermediate revisions not shown) | |||
| Line 97: | Line 97: | ||
| - | =='' | + | ==''Wednesday''== |
==='''Plan'''=== | ==='''Plan'''=== | ||
| Line 105: | Line 105: | ||
*'''We ran a low melting agarose gel 1 hour at 80 volts, then cut off the band from gel.''' | *'''We ran a low melting agarose gel 1 hour at 80 volts, then cut off the band from gel.''' | ||
| - | '''At Claudia's lab we tried three different | + | '''At Claudia's lab we tried three two different kits to recover DNA from gel.''' |
| - | *'''''' | + | *'''Mobio ultra clean Microbial DNA kit''' |
| + | *'''Axygen.....''' | ||
| + | *''' | ||
| - | '''The | + | '''The digestions were done with following amounts.''' |
{| border="1" | {| border="1" | ||
| Line 134: | Line 136: | ||
|30μl | |30μl | ||
|} | |} | ||
| - | |||
==''Thursday''== | ==''Thursday''== | ||
| Line 140: | Line 141: | ||
==='''Plan'''=== | ==='''Plan'''=== | ||
| - | '''We | + | '''We then proceed to diluted PCR fragments''' |
| - | '''For this we did the following | + | '''For this we did the following operations:''' |
'''Poner las fórmulas usadas en un formato bonito''' | '''Poner las fórmulas usadas en un formato bonito''' | ||
| - | '''We did the ligations using the following amounts | + | '''We did the ligations using the following amounts:''' |
{| border="1" | {| border="1" | ||
| Line 170: | Line 171: | ||
| - | '''We used the follow | + | '''We used the follow denomination.''' |
| - | *''' | + | *'''Band purified vector : Pu''' |
| - | *''' | + | *'''iGEM's vector (Vial with green cover): P''' |
| - | *''' | + | *'''Digested vector: S''' |
| - | '''After one hour | + | '''After one hour ligation we transformed by heat shock''' |
| - | '''and | + | '''and kept overnight the plates at 37ºC''' |
| Line 184: | Line 185: | ||
==''Friday''== | ==''Friday''== | ||
| - | '''Today we | + | '''Today we discussed what we going to do with the AFP (anti freeze protein),''' |
| - | '''We | + | '''We decided transform by heat shock and plating it on kanamicina medium, then prepare''' |
| - | '''miniprep and cut with Xba and | + | '''miniprep plasmid and cut it with ''Xba''I and ''Pst''I. We have to ligate it to the backbone J13002''' |
| - | '''the | + | '''the backbone had to be cut with ''Spe''I and ''Pst''I.''' |
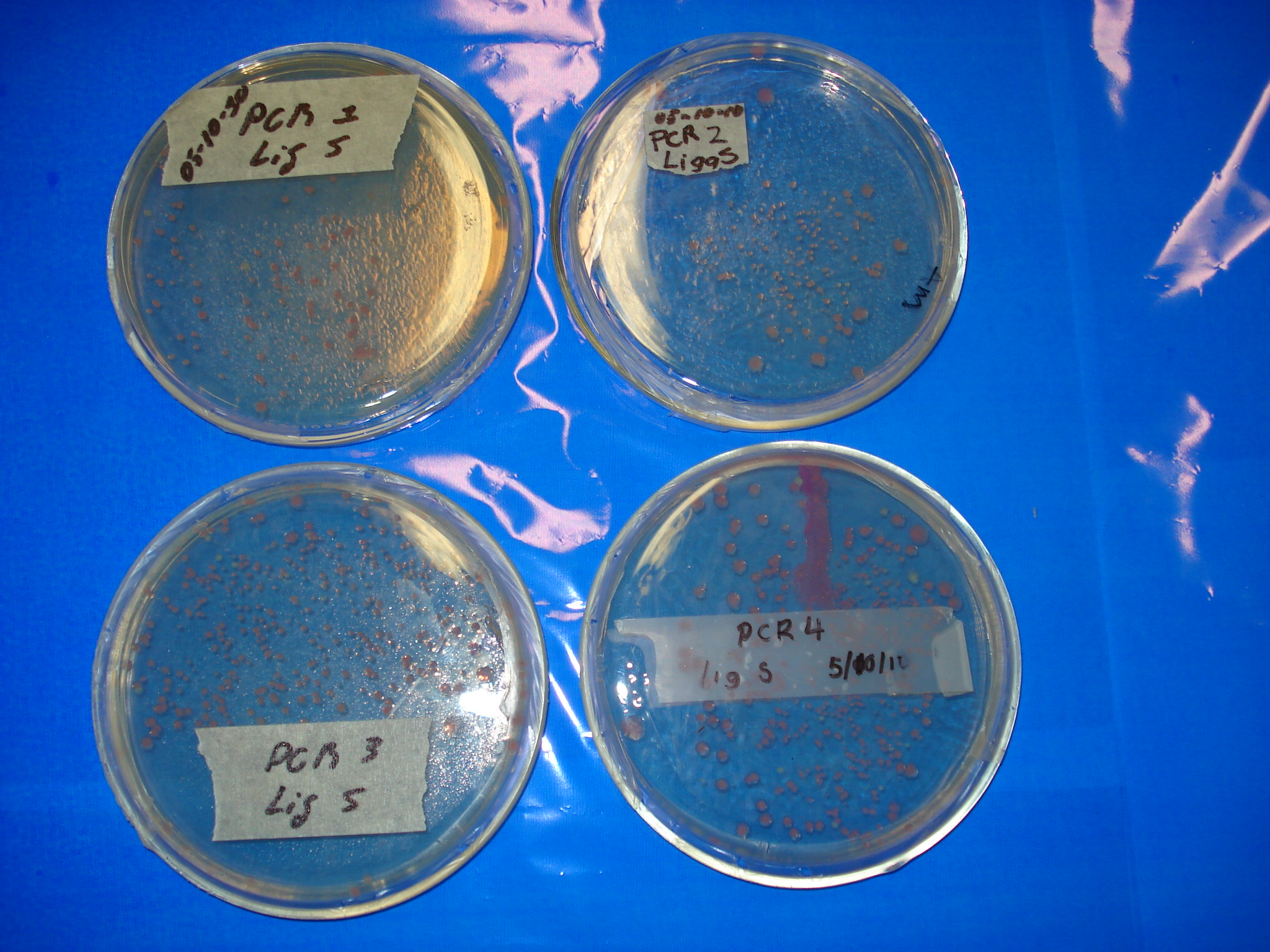
| - | '''Meanwhile | + | '''Meanwhile, here are the previous day ligations.''' |
| - | *'''PCR1 Lig S Positive''' | + | *'''PCR1 Lig S Positive!!!!!!!''' |
| - | *'''PCR2 Lig S Positive''' | + | *'''PCR2 Lig S Positive!!!!''' |
| - | *'''PCR3 Lig S Positive''' | + | *'''PCR3 Lig S Positive!!!!''' |
| - | *'''PCR4 Lig Pu Positive''' | + | *'''PCR4 Lig Pu Positive!!!!''' |
| - | *'''PCR4 Lig S Positive''' | + | *'''PCR4 Lig S Positive!!!!''' |
[[Image:Labo_254.JPG|400px]] | [[Image:Labo_254.JPG|400px]] | ||
Latest revision as of 06:41, 27 October 2010
Week #4
27th September - 01 October 2010
Monday
Well...we are stuck with the vector. This week we have to get it.
- We have achieved a nano spectrophotometer and readings are below:
| ng/μl | 260/280 | 260/230 | |
| PsB1C3 | 0.60 | -0.73 | -0.0 |
| PCR 1 red | 427.70 | 1.71 | 1.71 |
| PCR 2 red | 483.20 | 1.74 | 1.71 |
| PCR 3 red | 410.70 | 1.76 | 2.02 |
| PCR 4 red | 431.05 | 1.61 | 1.88 |
| PCR 1 blue | 533.35 | 1.71 | 1.75 |
| PCR 2 blue | 536.00 | 1.72 | 1.82 |
| PCR 3 blue | 577.50 | 1.69 | 1.59 |
| PCR 4 blue | 627.55 | 1.70 | 1.56 |
| PsB1C3 | 4.10 | 2.62 | 1.01 |
- Yep, our trouble is the miniprep method
we have a low concentration of vector plasmid.
We have to work an try to get more plasmid and used diluted PCR products
is not possible achieve ligations with this vector an insert ratio.
Our advisor proposed use low Melting agarose
to extract the plasmid from the gel.
Tuesday
We ran with a low meelting agarose and cut off the band
we precipitated using alchol and salts in order
to get enough vector to do the ligations.
Wednesday
Plan
- We set up all to do the ligations.
- We digested both vector and insert with EcoRI and PstI .
- We ran a low melting agarose gel 1 hour at 80 volts, then cut off the band from gel.
At Claudia's lab we tried three two different kits to recover DNA from gel.
- Mobio ultra clean Microbial DNA kit
- Axygen.....
The digestions were done with following amounts.
| DNA | 20μl |
| Buffer NB2 | 3μl |
| EcoRI | 2μl |
| PstI | 2μl |
| H2O | 2.4μl |
| BSA | 0.6μl |
| Total | 30μl |
Thursday
Plan
We then proceed to diluted PCR fragments
For this we did the following operations:
Poner las fórmulas usadas en un formato bonito
We did the ligations using the following amounts:
| Insert [20ng/μl] | 5μl |
| Vector | 1μl |
| Reaction buffer | 1μl |
| T4 DNA Ligase | 1μl |
| H2O | 2μl |
| Total | 10μl |
We used the follow denomination.
- Band purified vector : Pu
- iGEM's vector (Vial with green cover): P
- Digested vector: S
After one hour ligation we transformed by heat shock
and kept overnight the plates at 37ºC
Friday
Today we discussed what we going to do with the AFP (anti freeze protein),
We decided transform by heat shock and plating it on kanamicina medium, then prepare
miniprep plasmid and cut it with XbaI and PstI. We have to ligate it to the backbone J13002
the backbone had to be cut with SpeI and PstI.
Meanwhile, here are the previous day ligations.
- PCR1 Lig S Positive!!!!!!!
- PCR2 Lig S Positive!!!!
- PCR3 Lig S Positive!!!!
- PCR4 Lig Pu Positive!!!!
- PCR4 Lig S Positive!!!!
 "
"