Team:Mexico-UNAM-CINVESTAV/Modeling/Simulations
From 2010.igem.org
m (Con footer) |
|||
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Mexico-UNAM-CINVESTAV-HEADER}} | {{Mexico-UNAM-CINVESTAV-HEADER}} | ||
| + | == Cold shock response simulations == | ||
| + | Our model was implemented in the software tool CellDesigner 4.1. And the model file can be downloaded from [[https://2010.igem.org/Image:MEXICO_UNAM_CINVESTAV_CspA_system.zip here]] | ||
| - | [[Image:MEXICO_UNAM_CINVESTAV_Csp_sim1.jpg|thumb|center|500px| Simulation 1 of the short model of CSP response |Simulation | + | The estimation of the parameters has been a serious problem for the design of any model, and in our case we are not the exception. Unfortunately we were not able to obtain enough laboratory meditions and then our model is more an qualitative than quantitative approach. |
| + | |||
| + | The main behavior that we tried to reproduce was the induction of the expression of the CspA protein during cold shock and later the adapted state of the cell where the synthesis of the cold shock proteins is low and the rate of the synthesis of the rest of the proteins stabilizes to certain level. | ||
| + | |||
| + | |||
| + | |||
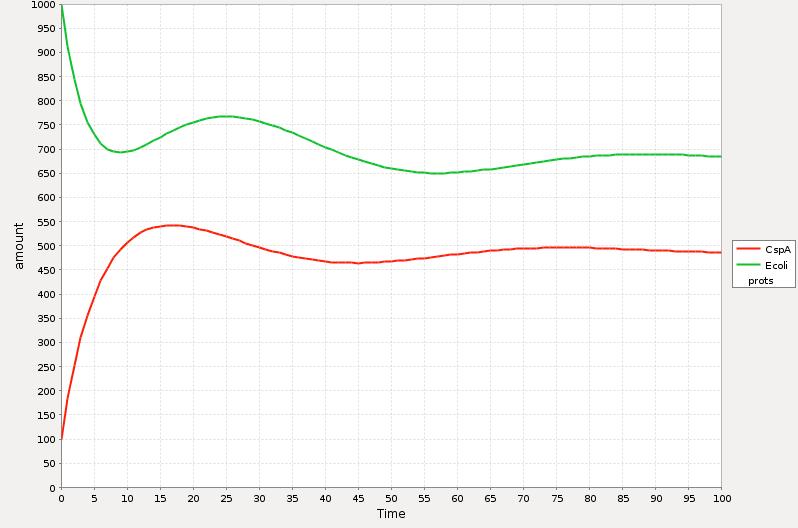
| + | [[Image:MEXICO_UNAM_CINVESTAV_Csp_sim1.jpg|thumb|center|500px| Simulation 1 of the short model of CSP response |Simulation 1]] | ||
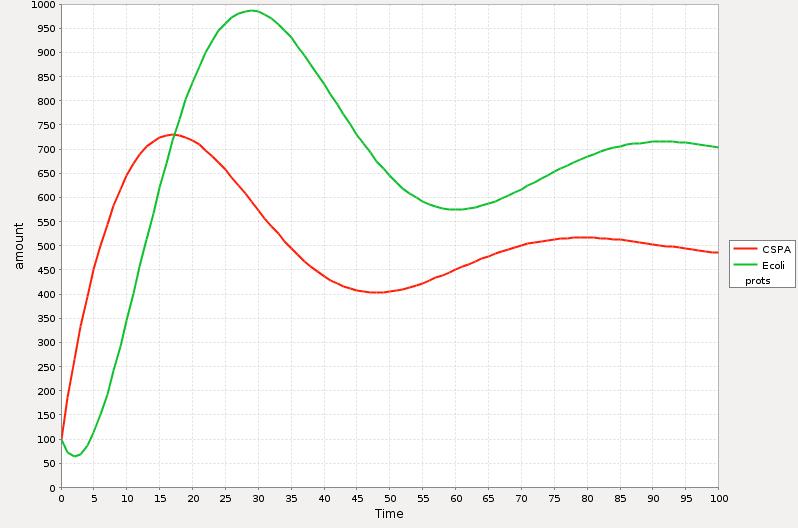
[[Image:MEXICO_UNAM_CINVESTAV_Csp_sim2.jpg|thumb|center|500px| Simulation 2 of the short model of CSP response |Simulation 2]] | [[Image:MEXICO_UNAM_CINVESTAV_Csp_sim2.jpg|thumb|center|500px| Simulation 2 of the short model of CSP response |Simulation 2]] | ||
| + | In the Simulations 1 and 2 is represented the qualitative response of the cold shock system in concentration during time. The time t=0 represents the exact point where the system is exposed to the temperature downshift. The level of the ''E. coli'' proteins decrease until enough CspA is present in the media and help them to recover certain level. The amount of CspA increases until ''E. coli'' proteins reach a maximum level and then decays to a stable one by the negative effect of the rest of the proteins. | ||
| + | |||
| + | The main difference between the response curve in the Theory section is that we are plotting here the total of cocentrations of the proteins during time and the response curse shows the change of the synthesis rate before, during and after cold shock. | ||
| + | |||
| + | |||
| + | As a further work we will introduce a more realistic parameters which will be determinated by experimental meditions. | ||
| + | |||
| + | |||
| + | |||
| + | == Visualization of the Antifreeze protein == | ||
| + | <html> | ||
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="425" height="349" src="http://www.youtube.com/embed/2S3dftpbTKU?rel=0" frameborder="0"></iframe> | ||
| + | |||
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="425" height="349" src="http://www.youtube.com/embed/Qb9KC8UulPs?rel=0" frameborder="0"></iframe> | ||
| + | |||
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="425" height="349" src="http://www.youtube.com/embed/DNLuuKBI0R4?rel=0" frameborder="0"></iframe> | ||
| + | |||
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="425" height="349" src="http://www.youtube.com/embed/YqT1ufeY2Xc?rel=0" frameborder="0"></iframe> | ||
| + | |||
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="425" height="349" src="http://www.youtube.com/embed/Fr03hNPt354?rel=0" frameborder="0"></iframe> | ||
| + | |||
| + | </html> | ||
| + | == Software references == | ||
| + | * Funahashi, A., Tanimura, N., Morohashi, M., and Kitano, H., CellDesigner: a process diagram editor for gene-regulatory and biochemical networks, BIOSILICO, 1:159-162, 2003 [doi:10.1016/S1478-5382(03)02370-9] | ||
| + | * Funahashi, A.; Matsuoka, Y.; Jouraku, A.; Morohashi, M.; Kikuchi, N.; Kitano, H."CellDesigner 3.5: A Versatile Modeling Tool for Biochemical Networks" Proceedings of the IEEE Volume 96, Issue 8, Aug. 2008 Page(s):1254 - 1265 [doi 10.1109/JPROC.2008.925458] | ||
Latest revision as of 20:10, 27 October 2010
Cold shock response simulations
Our model was implemented in the software tool CellDesigner 4.1. And the model file can be downloaded from [here]
The estimation of the parameters has been a serious problem for the design of any model, and in our case we are not the exception. Unfortunately we were not able to obtain enough laboratory meditions and then our model is more an qualitative than quantitative approach.
The main behavior that we tried to reproduce was the induction of the expression of the CspA protein during cold shock and later the adapted state of the cell where the synthesis of the cold shock proteins is low and the rate of the synthesis of the rest of the proteins stabilizes to certain level.
In the Simulations 1 and 2 is represented the qualitative response of the cold shock system in concentration during time. The time t=0 represents the exact point where the system is exposed to the temperature downshift. The level of the E. coli proteins decrease until enough CspA is present in the media and help them to recover certain level. The amount of CspA increases until E. coli proteins reach a maximum level and then decays to a stable one by the negative effect of the rest of the proteins.
The main difference between the response curve in the Theory section is that we are plotting here the total of cocentrations of the proteins during time and the response curse shows the change of the synthesis rate before, during and after cold shock.
As a further work we will introduce a more realistic parameters which will be determinated by experimental meditions.
Visualization of the Antifreeze protein
Software references
- Funahashi, A., Tanimura, N., Morohashi, M., and Kitano, H., CellDesigner: a process diagram editor for gene-regulatory and biochemical networks, BIOSILICO, 1:159-162, 2003 [doi:10.1016/S1478-5382(03)02370-9]
- Funahashi, A.; Matsuoka, Y.; Jouraku, A.; Morohashi, M.; Kikuchi, N.; Kitano, H."CellDesigner 3.5: A Versatile Modeling Tool for Biochemical Networks" Proceedings of the IEEE Volume 96, Issue 8, Aug. 2008 Page(s):1254 - 1265 [doi 10.1109/JPROC.2008.925458]
 |
 |
 |
 |
 "
"