Team:Lethbridge/Notebook/Lab Work/June
From 2010.igem.org
Adam.smith4 (Talk | contribs) (→June 2/2010) |
Adam.smith4 (Talk | contribs) (→June 2/2010) |
||
| Line 94: | Line 94: | ||
<b>Results:</b> <br> | <b>Results:</b> <br> | ||
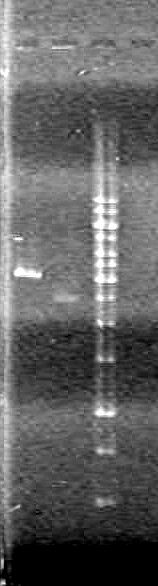
| - | [[image:100602 JV rbs-xylE.JPG|75px|none]] | + | [[image:100602 JV rbs-xylE.JPG|75px|none]]<br> |
| + | <b>Conclusions:</b> Plasmid DNA prep and restriction was successful.<br><br> | ||
| + | |||
| + | <b>Objective:</b> Ligate rbs-xylE (Bba_J33204) to our double terminator, and insert it into the pSB1T3 plasmid backbone.<br> | ||
| + | <b>Method:</b><br> | ||
| + | <ul> | ||
| + | <li><u>Restrictions</u><br> | ||
| + | *Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)<br> | ||
| + | *Restrict the double terminator with XbaI and PstI (Tango Buffer)<br> | ||
| + | *Restrict pSB1T3 with EcoRI and PstI (Red Buffer)<br> | ||
| + | Set up reactions as follows:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>15.5</td></tr> | ||
| + | <tr><td>Buffer</td><td>2</td></tr> | ||
| + | <tr><td>pDNA</td><td>2</td></tr> | ||
| + | <tr><td>Enzyme</td><td>0.25 + 0.25</td></tr></table> | ||
| + | |||
| + | Set up control reaction as follows: | ||
| + | *MilliQ H<sub>2</sub>O - 16µL<br> | ||
| + | *Buffer - 2µL<br> | ||
| + | *pDNA - 2µL<br> | ||
| + | Incubated reactions for 65 minutes at 37<sup>o</sup>C<br> | ||
| + | Killed enzymes by incubating reactions for 10 minutes at 65<sup>o</sup>C<br> | ||
| + | |||
| + | <li><u>Ligation</u><br> | ||
| + | Reaction set up as follows: | ||
| + | *T4 DNA ligase - 0.25µL<br> | ||
| + | *rbs-xylE - 5µL<br> | ||
| + | *dT - 3µL<br> | ||
| + | *pSB1T3 - 8µL<br> | ||
| + | *10x Ligation Buffer - 2µL<br> | ||
| + | *MilliQ H<sub>2</sub>O - 1.75µL<br> | ||
| + | Incubated reactions overnight at room temperature (total of 19.5 hours)<br> | ||
| + | Killed enzymes by incubating reactions for 10 minutes at 80<sup>o</sup>C<br> | ||
| + | </ul> | ||
===June 3/2010=== | ===June 3/2010=== | ||
<b>Objective:</b> Ligate pLacI to sRBS-Lum-dT using the three antibiotic assembly method (according to Tom Knight's protocol). Previous ligation had miniscule quantities of DNA in ligation, not surprising it didn't work.<br> | <b>Objective:</b> Ligate pLacI to sRBS-Lum-dT using the three antibiotic assembly method (according to Tom Knight's protocol). Previous ligation had miniscule quantities of DNA in ligation, not surprising it didn't work.<br> | ||
Revision as of 00:51, 11 June 2010

| 
| 
| ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
Contents |
June 2010
June 1/2010
JV quantified the amount of DNA in gels run to date using ImageJ software. Results to be posted in working plasmids box.
Objective: Transform plasmids into DH5α
Method: Follow competent cell transformation protocol to transform the following:
From our ligations:
- pLacI-sRBS-Lumazine-dT
- pLacI-sRBS-Lumazine-dT
- mms6 (A6)
- mms6 (B6)
- xylE (C4)
- xylE (B4)
From the 2010 Parts Distribution:
- ECFP (Bba_E0020)
- EYFP (Bba_E0030)
- BglII Endonuclease (Bba_K112106)
June 2/2010
(In Lab: JV)
Objective: Isolate plasmid DNA of RBS-xylE (BBa_J33204) from DH5α cells and confirm results.
Method: "Mini-prep" the plasmid DNA using boiling lysis miniprep. Then restrict the DNA once and run on a 1% agarose gel (TAE).
Restriction Reaction
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 15.75 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
| EcoRI | 0.25 |
Unrestricted Control
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 16 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
DNA was restricted for 80 minutes at 37oC.
Analyzed results on a 1% agarose gel. Load order as follows:
| Lane | Sample | Volume Sample (µL) | Volume Loading Dye (µL) |
| 1 | Restricted RBS-xylE | 10 | 2 |
| 1 | Unestricted RBS-xylE† | 1 | 2 |
| 1 | 1kb Ladder†† | 2 | 2 |
† Added 9µL MilliQ H2O
†† Added 8µL MilliQ H2O
Ran gel at 100V from 2 hours.
Results:
Conclusions: Plasmid DNA prep and restriction was successful.
Objective: Ligate rbs-xylE (Bba_J33204) to our double terminator, and insert it into the pSB1T3 plasmid backbone.
Method:
- Restrictions
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
- Restrict the double terminator with XbaI and PstI (Tango Buffer)
- Restrict pSB1T3 with EcoRI and PstI (Red Buffer)
Component Volume (µL) MilliQ H2O 15.5 Buffer 2 pDNA 2 Enzyme 0.25 + 0.25 Set up control reaction as follows:
- MilliQ H2O - 16µL
- Buffer - 2µL
- pDNA - 2µL
Incubated reactions for 65 minutes at 37oC
Killed enzymes by incubating reactions for 10 minutes at 65oC
- Ligation
Reaction set up as follows:- T4 DNA ligase - 0.25µL
- rbs-xylE - 5µL
- dT - 3µL
- pSB1T3 - 8µL
- 10x Ligation Buffer - 2µL
- MilliQ H2O - 1.75µL
Killed enzymes by incubating reactions for 10 minutes at 80oC
</ul>June 3/2010
Objective: Ligate pLacI to sRBS-Lum-dT using the three antibiotic assembly method (according to Tom Knight's protocol). Previous ligation had miniscule quantities of DNA in ligation, not surprising it didn't work. - Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
 "
"