Team:Lethbridge/Lab Work
From 2010.igem.org
Adam.smith4 (Talk | contribs) |
Adam.smith4 (Talk | contribs) |
||
| Line 1: | Line 1: | ||
Back to [[Team:Lethbridge/Notebook|Notebook]]<br> | Back to [[Team:Lethbridge/Notebook|Notebook]]<br> | ||
| - | + | ==April== | |
| + | ===April 13/2010=== | ||
| + | (In the Lab: JV, AS)<br> | ||
<b>Objective:</b> Test Restriction Endonucleases for Activity<br> | <b>Objective:</b> Test Restriction Endonucleases for Activity<br> | ||
<b>Relevant Information:</b><br> | <b>Relevant Information:</b><br> | ||
| Line 56: | Line 58: | ||
<b>Conclusion:</b> Will have to re-run experiment with DNA that is present at high enough concentrations to visualize by ethidium bromide staining<br><br><br> | <b>Conclusion:</b> Will have to re-run experiment with DNA that is present at high enough concentrations to visualize by ethidium bromide staining<br><br><br> | ||
| - | + | ==May== | |
| - | + | ===May 5/2010=== | |
| - | + | (in the lab: JV)<br> | |
<b>Objective:</b> Test Restriction Endonucleases for activity (take 2)<br> | <b>Objective:</b> Test Restriction Endonucleases for activity (take 2)<br> | ||
<b>Relevant Information:</b><br> | <b>Relevant Information:</b><br> | ||
| Line 117: | Line 119: | ||
<b>Conclusion:</b> Test other source of SpeI to see if it has any activity.<br><br> | <b>Conclusion:</b> Test other source of SpeI to see if it has any activity.<br><br> | ||
| - | + | ===May 6/2010=== | |
| + | (in the lab:KG, AS)<br> | ||
<b>Objective:</b> To check if the old SpeI enzyme (exp date: March 2011) will cleave plasmid DNA, since we believe the newer SpeI enzyme (exp date: 2012) does not.<br> | <b>Objective:</b> To check if the old SpeI enzyme (exp date: March 2011) will cleave plasmid DNA, since we believe the newer SpeI enzyme (exp date: 2012) does not.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 154: | Line 157: | ||
| - | + | ===May 10/2010=== | |
| + | (in the lab:JV)<br> | ||
<b>Objective:</b> To analyze the restriction test done by KG and AS on May 6/2010 by agarose electrophoresis<br><br> | <b>Objective:</b> To analyze the restriction test done by KG and AS on May 6/2010 by agarose electrophoresis<br><br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 174: | Line 178: | ||
It appears as though both SpeI enzymes are working properly here. We will utilize the newer batch of SpeI (expires 2012) from this point forward.<br><br> | It appears as though both SpeI enzymes are working properly here. We will utilize the newer batch of SpeI (expires 2012) from this point forward.<br><br> | ||
| - | + | <b>Objective:</b>Make 24 LB agar plates with 100µg/mL ampicillin antibiotic.(JV,KG,AV)<br> | |
| - | <b>Objective:</b>Make 24 LB agar plates with 100µg/mL ampicillin antibiotic.<br> | + | |
<b>Method:</b>Make 2L of LB media with agar<br> | <b>Method:</b>Make 2L of LB media with agar<br> | ||
2x10g Tryptone<br> | 2x10g Tryptone<br> | ||
| Line 185: | Line 188: | ||
Have 4x500mL of LB with Agar<br> | Have 4x500mL of LB with Agar<br> | ||
Add 500µL of stock ampicillin to 500mL of media<br><br> | Add 500µL of stock ampicillin to 500mL of media<br><br> | ||
| - | + | ||
| + | ===May 11/2010 Evening=== | ||
| + | (in the lab: KG, AV, MC, TF, JV, JS)</b> | ||
<b>Objective:</b> To transform the following plasmids into DH5α <i>E.coli</i> cells. | <b>Objective:</b> To transform the following plasmids into DH5α <i>E.coli</i> cells. | ||
<table><table border="3"> | <table><table border="3"> | ||
| Line 224: | Line 229: | ||
<li>pTet</li></ul><br> | <li>pTet</li></ul><br> | ||
| - | + | ===May 12/2010=== | |
| + | (in the lab: JV)<br> | ||
<b>Objective: Miniprep of plasmid DNA from transformed cells</b>(JV, AV, HB)<br> | <b>Objective: Miniprep of plasmid DNA from transformed cells</b>(JV, AV, HB)<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 343: | Line 349: | ||
Start overnight cultures of cells that grew for plasmid prep and sequencing.<br><br> | Start overnight cultures of cells that grew for plasmid prep and sequencing.<br><br> | ||
| - | + | ===May 14/2010=== | |
| + | (in the lab: JV)<br> | ||
<b>Objective:</b> Quantify pDNA concentration in order to ensure sufficient material for sequence analysis.<br> | <b>Objective:</b> Quantify pDNA concentration in order to ensure sufficient material for sequence analysis.<br> | ||
<b>Method:</b> Measure absorbance of samples at 260nm.<br> | <b>Method:</b> Measure absorbance of samples at 260nm.<br> | ||
| Line 414: | Line 421: | ||
There is plasmid DNA in each sample which, when cut with both the prefix and suffix enzyme, yields a band approximately 2000bp (size of pSB1A3 is 2157bp).<br><br> | There is plasmid DNA in each sample which, when cut with both the prefix and suffix enzyme, yields a band approximately 2000bp (size of pSB1A3 is 2157bp).<br><br> | ||
| - | + | ===May 17/2010=== | |
| + | (in the lab: JV, AV)<br> | ||
Make agar plates with 100µg/mL of ampicillin<br> | Make agar plates with 100µg/mL of ampicillin<br> | ||
Make 5 x 5mL sterile liquid SOC broth<br> | Make 5 x 5mL sterile liquid SOC broth<br> | ||
Make 13 x 5mL sterile liquid LB broth<br><br> | Make 13 x 5mL sterile liquid LB broth<br><br> | ||
| - | + | ===May 17/2010 Evening=== | |
| + | (in the lab: TF, AS)<br> | ||
<b>Objective:</b> To grow cells for future use<br> | <b>Objective:</b> To grow cells for future use<br> | ||
Streaked plates from glycerol stocks of the following:<br> | Streaked plates from glycerol stocks of the following:<br> | ||
| Line 451: | Line 460: | ||
Only sRBS (D5) and pBad-TetR cells (from transformation plates) grew.<br> | Only sRBS (D5) and pBad-TetR cells (from transformation plates) grew.<br> | ||
| - | + | ===May 18/2010=== | |
| + | (in the lab: JV, AV, HB)<br> | ||
<b>Objective:</b> To isolate plasmid DNA of pBad-TetR and sRBS (D5), using [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]].\<br><br> | <b>Objective:</b> To isolate plasmid DNA of pBad-TetR and sRBS (D5), using [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]].\<br><br> | ||
Revision as of 00:15, 22 May 2010
Back to Notebook
Contents |
April
April 13/2010
(In the Lab: JV, AS)
Objective: Test Restriction Endonucleases for Activity
Relevant Information:
Endonucleases available
| Endonuclease | Optimal Buffer** | Other Buffers |
| EcoRV | None | 2xT(100%); O,G(50-100%) |
| EcoRI | Red | O(100%);R(100%)*;2xT(100%) |
| BcuI/SpeI | Tango | B(50-100%);G(50-100%) |
| XbaI | Tango | B,G,2xT(50-100%) |
| PstI | Orange | R(100%); B,G,T,2xT(50-100%) |
| DpnI | Tango | B,G(100%): O,R,2xT(50-100%) |
- Star Activity
- Optimal Buffer from Fermentas
- Optimal Buffer from Fermentas
Use pUC19 plasmid as test, it has cut sites for EcoRI, PstI, XbaI (unsure about BcuI/SpeI, DpnI but will try anyways), and none for EcoRV
Red Buffer: EcoRI, PstI, Control (No Enzyme)
Tango Buffer: BcuI/SpeI, XbaI, DpnI, Control (No Enzyme>
Methods:
Set up Master Mixes:
| Red MM | per tube (µL) | Total (µL) |
| MilliQ H20 | 13.75 | 55 |
| Red Buffer (10x) | 2 | 7 |
| pUC19 (10pg/µL) | 2 | 7 |
| Total | 19.75 | 69 |
| Tango MM | per tube (µL) | Total (µL) |
| MilliQ H20 | 13.75 | 55 |
| Tango Buffer (10x) | 2 | 7 |
| pUC19 (10pg/µL) | 2 | 7 |
| Total | 19.75 | 69 |
To each tube, add 19.75µL of master mix and 0.25µL of enzyme
Incubated reaction mixes at 37oC (Start:7:00pm; End:7:45pm)
Add 3.3µL of 6x loading dye to each reaction mixture and load 10µL final volume onto a 1% agarose (in TAE) gel.
Add 1µL of 6x loading dye to 1µL of GeneRuler 1kb ladder (at 0.5µg/µL)
Gel loading order as follows:
| Lane | Sample |
| 1 | 1kb Ladder |
| 2 | Tango Control |
| 3 | DpnI (Tango) |
| 4 | BcuI/SpeI (Tango) |
| 5 | XbaI (Tango) |
| 6 | EcoRI (Red) |
| 7 | PstI (Red) |
| 8 | Red Control |
| 9 | Empty |
| 10 | Empty |
Ran gel at 100V for 1 hour
Results: pUC19 plasmid DNA not present at a high enough concentration to visualize by ethidium bromide staining (1kb ladder did stain).
Conclusion: Will have to re-run experiment with DNA that is present at high enough concentrations to visualize by ethidium bromide staining
May
May 5/2010
(in the lab: JV)
Objective: Test Restriction Endonucleases for activity (take 2)
Relevant Information:
Plasmid DNA used here will be "ES-pSB-CEYFP" from last year's plasmid stocks
Prefix Enzymes are: EcoRI and XbaI
Suffix Enyzmes are: SpeI and PstI
(JV worked out in lab notebook which buffers would be best for each prefix/suffix enzyme combination)
Reactions will be assembled as follows:
| Enzyme</td> | Buffer</td> | Volume MM(µL)</td> | Volume Enzyme(µL)</td></tr> |
| PstI | Red | 19.75 | .25 |
| XbaI | Tango | 19.75 | .25 |
| SpeI | Tango | 19.75 | .25 |
| EcoRI | Red | 19.75 | .25 |
| EcoRI/SpeI | Red | 19.5 | .25+.25 |
| XbaI/SpeI | Tango | 19.5 | .25+.25 |
| EcoRI/PstI | Red | 19.5 | .25+.25 |
| XbaI/PstI | Tango | 19.5 | .25+.25 |
Make up Master Mixes as follows:
| Red MM</td> | per tube(µL)</td> | Total*(µL)</td></tr> |
| MilliQ H20 | 15.75 | 86.675 |
| Red Buffer (10x) | 2 | 11 |
| pDNA** | 2 | 11 |
| Tango MM</td> | per tube(µL)</td> | Total*(µL)</td></tr> |
| MilliQ H20 | 15.75 | 86.675 |
| Tango Buffer (10x) | 2 | 11 |
| pDNA** | 2 | 11 |
- Volume per reaction multiplied by 5.5
- Unknown concentration of pDNA
- Unknown concentration of pDNA
Incubated for 70min at 37oC (Start-1:05pm; End-2:15pm)
Added 3.3µL of 6x loading dye to each reaction mixture and loaded 10µL onto a 1% agarose gel (in TAE)
Added 1µL of 6x loading dye to 2µL of gene ruler 1kb ladder
Load order as follows:
| Lane</td> | Sample</td> | Volume Loaded (µL)</td></tr> |
| 1 | pSB-CEYFP/PstI | 10 |
| 2 | pSB-CEYFP/EcoRI | 10 |
| 3 | pSB-CEYFP/EcoRI/PstI | 10 |
| 4 | pSB-CEYFP/EcoRI/SpeI | 10 |
| 5 | pSB-CEYFP/XbaI/PstI | 10 |
| 6 | pSB-CEYFP/XbaI | 10 |
| 7 | pSB-CEYFP/SpeI | 10 |
| 8 | pSB-CEYFP/XbaI/SpeI | 10 |
| 9 | pSB-CEYFP/Red Master Mix Control | 10 |
| 10 | pSB-CEYFP/Tango Master Mix Control | 10 |
| 11 | pSB-CEYFP/MilliQ H20 Control | 10 |
| 12 | Ladder | 4 |
Ran gel at 100V for 1 hour
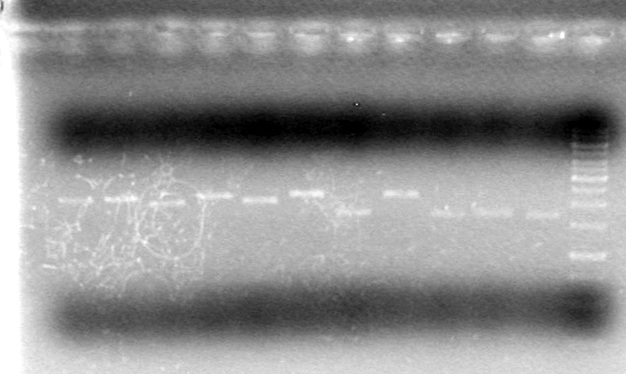
Results:
This gel shows that SpeI does not cut on its own, and does not cut when combined with other enzymes
Conclusion: Test other source of SpeI to see if it has any activity.
May 6/2010
(in the lab:KG, AS)
Objective: To check if the old SpeI enzyme (exp date: March 2011) will cleave plasmid DNA, since we believe the newer SpeI enzyme (exp date: 2012) does not.
Method:
| Red Master Mix | per tube (µL) | Total Volume* |
| MilliQ H20 Water | 15.75 | 63 |
| Red Buffer (10x) | 2 | 8 |
| pDNA** | 2 | 8 |
- Volume per tube multiplied by 4
- Used pSB NEYFP pDNA from cell E5 in plasmid box
- Used pSB NEYFP pDNA from cell E5 in plasmid box
Enzymes that will use Red Master Mix are: EcoRI+SpeI (old), EcoRI+SpeI (new)
Add 0.25µL of each enzyme to 19.5µL of master mix
| Tango Master Mix | per tube (µL) | Total Volume* |
| MilliQ H20 Water | 15.75 | 94.5 |
| Tango Buffer (10x) | 2 | 12 |
| pDNA** | 2 | 12 |
- Volume per tube multiplied by 6
- Used pSB NEYFP pDNA from cell E5 in plasmid box
- Used pSB NEYFP pDNA from cell E5 in plasmid box
Enzymes that will use Tango Master Mix are: SpeI (old), SpeI (new), XbaI+SpeI (old), XbaI+SpeI (new)
Add 0.25µL of each enzyme to 19.5µL of master mix
Incubated all reactions at 37oC for 1h (Start-8:30pm; End-9:30pm)
Will not be able to run on agarose gel tonight, will label them so JV can run them in the morning
Tube Names:
Master Mix 1 Control (Red Buffer)
Master Mix 2 Control (Tango Buffer)
E+S(N); EcoRI + SpeI(N)
E+S(O); EcoRI + SpeI(O)
X+S(N); XbaI + SpeI(N)
X+S(O); XbaI + SpeI(O)
S(N); SpeI(N)
S(O); SpeI(O)
Placed in -20oC freezer of later analysis by agarose electrophoresis
May 10/2010
(in the lab:JV)
Objective: To analyze the restriction test done by KG and AS on May 6/2010 by agarose electrophoresis
Method:
| Lane | Sample | Quantity Loaded (µL) |
| 1 | MM1 Control | 10 |
| 2 | MM2 Control | 10 |
| 3 | EcoRI+SpeI(N) | 10 |
| 4 | EcoRI+SpeI(O) | 10 |
| 5 | SpeI(N) | 10 |
| 6 | SpeI(O) | 10 |
| 7 | XbaI+SpeI(N) | 10 |
| 8 | XbaI+SpeI(O) | 10 |
| 9 | 1kb Ladder | 5 |
Run gel for 60min at 100V
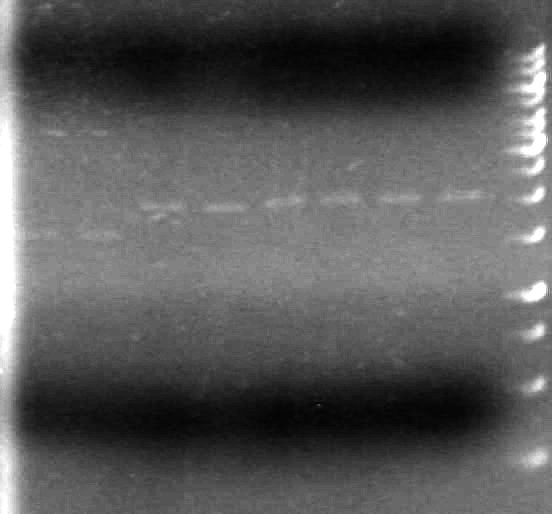
Results:
It appears as though both SpeI enzymes are working properly here. We will utilize the newer batch of SpeI (expires 2012) from this point forward.
Objective:Make 24 LB agar plates with 100µg/mL ampicillin antibiotic.(JV,KG,AV)
Method:Make 2L of LB media with agar
2x10g Tryptone
2X2.5g Yeast Extract
2x5g NaCl
2x10g Agar
Continued May 11/2010
(Stock Ampicillin solution is 100mg/mL)
Have 4x500mL of LB with Agar
Add 500µL of stock ampicillin to 500mL of media
May 11/2010 Evening
(in the lab: KG, AV, MC, TF, JV, JS)</b> Objective: To transform the following plasmids into DH5α E.coli cells.
| Construct Name (2009)</td> | Construct Location (2009) |
| Lumazine | J4 |
| Lumazine-dT | J5,J6 |
| sRBS-Lumazine-dT | J7,J8 |
| pBAD-TetR | I4 |
| pBAD | A5,F10 |
| sRBS | D5,E10 |
| pSB-CEYFP | E5,D6 |
| pSB-NEYFP | F5,C6 |
| C-term Tag | C10 |
| N-term Tag | D9,D10 |
| pTet | E4 |
| EYFP | A4 |
| CFP Complete | D4 |
Method: Followed Competent Cell Transformation protocol in Common Protocols section and plated on LB agar supplemented with ampicillin.
Results: The following plasmids were successfully transformed and formed colonies:
- Lumazine (J4)
- sRBS-Lumazine-dT (J7)
- sRBS-Lumazine-dT (J8)
- pBAD (A5)
- pBAD (F10)
- pSB-CEYFP
- pSB-NEYFP
- N-term tag
- EYFP (A4)
- CFP Complete (D4)
Conclusion: Need another attempt to transform the following plasmids:
- Lumazine-dT (J5,J6)
- pBAD-TetR
- sRBS (D5,E10)
- C-Term tag
- pTet
May 12/2010
(in the lab: JV)
Objective: Miniprep of plasmid DNA from transformed cells(JV, AV, HB)
Method:
- Inoculate 5mL of LB liquid media (with 100µL/mL Ampicillin) with cells from competent cells plates (picked with sterile toothpick).
- Allow cells in liquid culture to grow overnight in 37oC shaking incubator (300RPM) Purify plasmid DNA from cells by using "Boiling Lysis Plasmid Preparation" protocol in Common Protocols Section.
- CHANGE: Step 14, used MilliQ H2O (with 20ng/µL RNase A) instead of TE buffer.
Plasmids were transferred to the "iGEM 2010 - Working Plasmid DNA" box in the -20oC freezer in the iGEM lab. Plasmids were placed in the following cells:
| Construct | Cell in Working Plasmid Box (2010) | Original Cell in Old Box |
| sRBS-Lumazine-dT | A1 | J7 |
| sRNS-Lumazine-dT | A2 | J8 |
| CFP Complete | B6 | D4 |
| Lumazine | A3 | J4 |
| pBAD | A4 | A5 |
| pBAD | A5 | F10 |
| pSB-CEYFP | B5 | |
| pSB-NEYFP | B4 | |
| EYFP | B1 | A4 |
| N-term tag | B2 |
Also generated sterile glycerol stocks and placed in -80oC freezer in the 2010 iGEM box as follows:
| Construct | Cell Working Glycerol Stock Box (2010) |
| sRBS-Lumazine-dT (J7) | B2,C4,D2 |
| sRNS-Lumazine-dT (J8) | C6 |
| CFP Complete | A10, C8 |
| Lumazine | A8,B10 |
| pBAD (from A5) | B5,B9 |
| pBAD (from F10) | B3,B7 |
| pSB-CEYFP | C3,B5 |
| pSB-NEYFP | B6,C1 |
| EYFP | C7,B8 |
| N-term tag | C2,D4 |
Objective: Restrict plasmid DNA with restriction endonucleases (JV)
Method:
Have:
10 lanes of restricted plasmid DNA
10 lanes of unrestricted plasmid DNA
1 lane of buffer control
Use EcoRI (prefix cutter) and PstI (suffix cutter)
Pipetting Scheme for Restriction Tubes:
| Ingredient | Volume/tube (µL) | Total Volume* |
| MilliQ H2O | 15.5 | 155 |
| Red Buffer (10X) | 2 | 20 |
| EcoRI | 0.25 | 2.5 |
| PstI | 0.25 | 2.5 |
- Amount per tube multiplied by 10
Pipetting Scheme for Unrestricted reactions:
| Ingredient | Volume/tube (µL) | Total Volume* |
| MilliQ H2O | 16 | 160 |
| Red Buffer (10X) | 2 | 20 |
- Amount per tube multiplied by 10
Buffer Control will be 18µL MilliQ H2O + 2µL 10x Red Buffer.
Place in 37oC water bath at 2:55pm and removed at 4:57pm for a 2 hour incubation.
Analyzed restriction digests on a 1% agarose gel (large gel apparatus ~70mL)
Added 1µL of 6x DNA loading dye to 5µL of sample
Added 2µL of 6x DNA loading dye to 6µL of TAE buffer and 2µL of 1kb DNA mass ladder.
Loaded samples as follows:
| Lane</td> | Sample</td> | Volume Loaded (µL) |
| 1 | 1 kb Ladder | 5 |
| 2 | Buffer Control | 5 |
| 3 | pSB-NEYFP | 5 |
| 4 | Restricted Lumazine | 5 |
| 5 | Lumazine | 5 |
| 6 | Restricted pSB-NEYFP | 5 |
| 7 | pSB-CEYFP | 5 |
| 8 | Restricted pSB-CEYFP | 5 |
| 9 | pBAD | 5 |
| 10 | Restricted pBAD | 5 |
| 11 | EYFP | 5 |
| 12 | Restricted EYFP | 5 |
| 13 | CFP Complete | 5 |
| 14 | Restricted CFP Complete | 5 |
| 15 | sRBS-Lumazine-dT (J7) | 5 |
| 16 | Restricted sRBS-Lumazine-dT (J7) | 5 |
| 17 | N-term Tag | 5 |
| 18 | Restricted N-term Tag | 5 |
| 19 | sRBS-Lumazine-dT (J8) | 5 |
| 20 | Restricted sRBS-Lumazine-dT (J8) | 5 |
Ran gel at 100V for 90 minutes (Start-9:50pm; End-11:20pm)
Stained with ethidium bromide for 20 minutes.
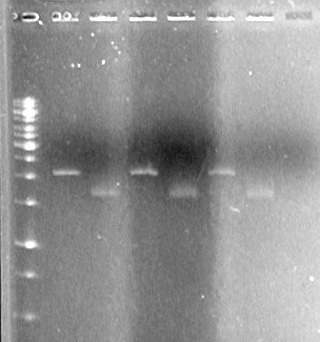
Results:
May 13/2010 Evening(in lab: AS,TF,KG,JS,MC)
Objective: To make a second attempt at transforming plasmids that didn't transform the first time. These plasmids are:
- Lumazine-dT (J5,J6)
- pBad-TetR
- sRBS (D5,E10)
- C-term tag
- pTet
All DH5α cells were used up in the last transformation, had to aliquot an additional 50x 20µL aliquots (MC,TF)
Transform plasmid DNA (Using "Competent Cell Transformation" Protocol) into newly aliquotted DH5α cells. (KG,JS)
NOTES:
AS concerned that there is something not quite right with LB liquid media added to transformed cells, but continued anyways (JV informed AS the next day that the LB liquid media had not been sterilized).
Plated all 250µL of culture.
Results:
| Construct</td> | Result |
| Lumazine-dT(1) | Growth present |
| sRBS-Lumazine-dT | Growth present |
| sRBS (D5) | Growth present |
| sRBS (E10) | Growth present |
| C-term tag | No growth present |
| pTet | No growth present |
Next Steps:
Make another attempt to transform the C-term tag and pTet constructs.
Start overnight cultures of cells that grew for plasmid prep and sequencing.
May 14/2010
(in the lab: JV)
Objective: Quantify pDNA concentration in order to ensure sufficient material for sequence analysis.
Method: Measure absorbance of samples at 260nm.
Results:
| Sample</td> | Absorbance at 260nm |
| sRBS-Lumazine-dT (J7) | 0.311 |
| sRBS-Lumazine-dT (J8) | 0.309 |
| CFP complete | 0.316 |
| N-term tag | 0.290 |
| pSB-CEYFP | 0.338 |
| pSB-NEYFP | 0.403 |
| pBAD (A5) | 0.282 |
| pBAD (F10) | 0.562 |
| EYFP | 0.389 |
| Lumazine | 0.221 |
Conclusion: All plasmids present in sufficient concentrations for sequence analysis.
Objective: Purify plasmid DNA from cells recently transformed.
Method:
- Inoculate 5mL of sterile LB liquid media (with 100µg/mL ampicillin) with cells picked from colonies of transformation plates, including the following:
Lumazine-dT (J5)
pBad-TetR
sRBS (D5,E10) - NOTE: Lumazine-dT did NOT grow overnight
- Followed "Boiling Lysis Plasmid Preparation (Miniprep)" protocol. (May 15/2010; JV,TF)
NOTE: Added 50µL of MilliQ H2O (with RNase A at a concentration of 20ng/µL) to dissolve pDNA instead of TE buffer.
Objective: Perform restriction digest on the above prepared plasmid DNA.
Method:
Used EcoRI as prefix cutter and PstI as suffix cutter.
Pipetting Scheme for Restriction Tubes:
| Ingredient | Volume/tube (µL) | Total Volume* |
| MilliQ H2O | 16 | 56 |
| Red Buffer (10X) | 2 | 7 |
| EcoRI | 0.25 | 0.875 |
| PstI | 0.25 | 0.875 |
- Amount per tube multiplied by 3.5
Add 18µL master mix to each plasmid DNA sample
Pipetting Scheme for Unrestricted reactions:
| Ingredient | Volume/tube (µL) | Total Volume* |
| MilliQ H2O | 16 | 56 |
| Red Buffer (10X) | 2 | 7 |
- Amount per tube multiplied by 3.5
Add 18µL master mix to each plasmid DNA sample
Buffer Control will be 18µL MilliQ H2O + 2µL 10x Red Buffer.
Place in 37oC water bath at 12:37pm and removed at 1:55pm for approximately 1 hour incubation.
Analyze samples on a 1% agarose gel (small gel apparatus).
Add 3.3µL of 6x DNA loading dye to each reaction mixture and load.
| Lane</td> | Sample</td> | Volume Loaded (µL)</td> |
| 1 | 1 kb Ladder | 4 |
| 2 | Restricted sRBS (E10) | 10 |
| 3 | sRBS (E10) | 10 |
| 4 | Restricted sRBS (D5) | 10 |
| 5 | sRBS (D5) | 10 |
| 6 | Restricted sRBS-Lumazine-dT | 10 |
| 7 | sRBS-Lumazine-dT | 10 |
| 8 | Red Buffer Control | 10 |
Ran gel at 100V for 75 minutes (Start-2:30pm; End-3:45pm)
Stained in ethidium bromide for 10 minutes
Results:
There is plasmid DNA in each sample which, when cut with both the prefix and suffix enzyme, yields a band approximately 2000bp (size of pSB1A3 is 2157bp).
May 17/2010
(in the lab: JV, AV)
Make agar plates with 100µg/mL of ampicillin
Make 5 x 5mL sterile liquid SOC broth
Make 13 x 5mL sterile liquid LB broth
May 17/2010 Evening
(in the lab: TF, AS)
Objective: To grow cells for future use
Streaked plates from glycerol stocks of the following:
iGEM 2007 -80oC Freezer Box:
| Construct | Cell type | Location of cells |
| xylE | DH5α | C4 |
| xylE | BL21(DE3) | B4 |
| C-term Bba | DH5α | H4 |
| C-term Bba | DH5α | I4 |
| C-term Bba | DH5α | J4 |
| Mr. Gene mms6 | DH5α | A6 |
| Mr. Gene mms6 | DH5α | B6 |
iGEM 2010 -80oC Freezer Box:
| Construct | Cell type | Location of cells |
| pLacI | DH5α | B1 |
| dT | BL21(DE3) | D1 |
Incubated at 37oC, beginning at 20h00 (8:00pm)
Made liquid cultures from cells taken from transformation plates (grown on May 13/2010):
- sRBS (D5)
- sRNB (E10)
- Lumazine-dT (1)
- pBad-TetR
Incubated at 37oC, beginning at 20h30 (8:30pm); shaking at 300RPM
Results:
All streak plates (with cells from glycerol stocks) grew.
Only sRBS (D5) and pBad-TetR cells (from transformation plates) grew.
May 18/2010
(in the lab: JV, AV, HB)
Objective: To isolate plasmid DNA of pBad-TetR and sRBS (D5), using boiling lysis miniprep.\
NOTE: Cells from liquid cultures grown last night (May 17/2010) were made into glycerol stocks and placed into the working glycerol stock box as follows:
- pBAD-TetR - E5
- pBAD-TetR - E6
- sRBS (D5) - E7
- sRBS (D5) - E8
 "
"