Team:Lethbridge/Lab Work
From 2010.igem.org
Adam.smith4 (Talk | contribs) (New page: This is our lab work page) |
Adam.smith4 (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| - | + | <b><font size=+2>April 13/2010</font> (In the Lab: JV, AS)</b><br> | |
| + | <b>Objective:</b> Test Restriction Endonucleases for Activity<br> | ||
| + | <b>Relevant Information:</b><br> | ||
| + | Endonucleases available | ||
| + | <table><table border="3"> | ||
| + | <tr><td>Endonuclease</td><td>Optimal Buffer**</td><td>Other Buffers</td></tr> | ||
| + | <tr><td>EcoRV</td><td>None</td><td>2xT(100%); O,G(50-100%)</td></tr> | ||
| + | <tr><td>EcoRI</td><td>Red</td><td>O(100%);R(100%)*;2xT(100%)</td></tr> | ||
| + | <tr><td>BcuI/SpeI</td><td>Tango</td><td>B(50-100%);G(50-100%)</td></tr> | ||
| + | <tr><td>XbaI</td><td>Tango</td><td>B,G,2xT(50-100%)</td></tr> | ||
| + | <tr><td>PstI</td><td>Orange</td><td>R(100%); B,G,T,2xT(50-100%)</td></tr> | ||
| + | <tr><td>DpnI</td><td>Tango</td><td>B,G(100%): O,R,2xT(50-100%)</td></tr> | ||
| + | </table> | ||
| + | *Star Activity<br> | ||
| + | **Optimal Buffer from Fermentas<br><br> | ||
| + | Use pUC19 plasmid as test, it has cut sites for EcoRI, PstI, XbaI (unsure about BcuI/SpeI, DpnI but will try anyways), and none for EcoRV <br> | ||
| + | <u>Red Buffer:</u> EcoRI, PstI, Control (No Enzyme)<br> | ||
| + | <u>Tango Buffer:</u> BcuI/SpeI, XbaI, DpnI, Control (No Enzyme><br><br> | ||
| + | <u>Methods:</u> | ||
| + | Set up Master Mixes: | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Red MM</b></td><td>per tube (µL)</td><td>Total (µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0</td><td>13.75</td><td>55</td></tr> | ||
| + | <tr><td>Red Buffer (10x)</td><td>2</td><td>7</td></tr> | ||
| + | <tr><td>pUC19 (10pg/µL)</td><td>2</td><td>7</td></tr> | ||
| + | <tr><td><b>Total</b></td><td>19.75</td><td>69</td></tr> | ||
| + | </table><br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Tango MM</b></td><td>per tube (µL)</td><td>Total (µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0</td><td>13.75</td><td>55</td></tr> | ||
| + | <tr><td>Tango Buffer (10x)</td><td>2</td><td>7</td></tr> | ||
| + | <tr><td>pUC19 (10pg/µL)</td><td>2</td><td>7</td></tr> | ||
| + | <tr><td><b>Total</b></td><td>19.75</td><td>69</td></tr> | ||
| + | </table><br> | ||
| + | To each tube, add <b>19.75µL</b> of master mix and <b>0.25µL</b> of enzyme<br> | ||
| + | Incubated reaction mixes at 37<sup>o</sup>C (Start:7:00pm; End:7:45pm)<br> | ||
| + | Add 3.3µL of 6x loading dye to each reaction mixture and load 10µL final volume onto a 1% agarose (in TAE) gel.<br> | ||
| + | Add 1µL of 6x loading dye to 1µL of GeneRuler 1kb ladder (at 0.5µg/µL)<br> | ||
| + | Gel loading order as follows:<br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb Ladder</td></tr> | ||
| + | <tr><td>2</td><td>Tango Control</td></tr> | ||
| + | <tr><td>3</td><td>DpnI (Tango)</td></tr> | ||
| + | <tr><td>4</td><td>BcuI/SpeI (Tango)</td></tr> | ||
| + | <tr><td>5</td><td>XbaI (Tango)</td></tr> | ||
| + | <tr><td>6</td><td>EcoRI (Red)</td></tr> | ||
| + | <tr><td>7</td><td>PstI (Red)</td></tr> | ||
| + | <tr><td>8</td><td>Red Control</td></tr> | ||
| + | <tr><td>9</td><td>Empty</td></tr> | ||
| + | <tr><td>10</td><td>Empty</td></tr> | ||
| + | </table><br> | ||
| + | Ran gel at 100V for 1 hour<br> | ||
| + | <b>Results:</b> pUC19 plasmid DNA not present at a high enough concentration to visualize by ethidium bromide staining (1kb ladder did stain). <br> | ||
| + | <b>Conclusion:</b> Will have to re-run experiment with DNA that is present at high enough concentrations to visualize by ethidium bromide staining<br><br><br> | ||
| + | |||
| + | |||
| + | <a name="may"></a> | ||
| + | <b><font size=+2>May 5/2010</font>(in the lab: JV)</b><br> | ||
| + | <b>Objective:</b> Test Restriction Endonucleases for activity (take 2)<br> | ||
| + | <b>Relevant Information:</b><br> | ||
| + | Plasmid DNA used here will be "ES-pSB-CEYFP" from last year's plasmid stocks<br> | ||
| + | Prefix Enzymes are: EcoRI and XbaI<br> | ||
| + | Suffix Enyzmes are: SpeI and PstI<br> | ||
| + | (JV worked out in lab notebook which buffers would be best for each prefix/suffix enzyme combination)<br> | ||
| + | Reactions will be assembled as follows:<br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Enzyme</td><td>Buffer</td><td>Volume MM(µL)</td><td>Volume Enzyme(µL)</td></tr></b> | ||
| + | <tr><td>PstI</td><td>Red</td><td>19.75</td><td>.25</td></tr> | ||
| + | <tr><td>XbaI</td><td>Tango</td><td>19.75</td><td>.25</td></tr> | ||
| + | <tr><td>SpeI</td><td>Tango</td><td>19.75</td><td>.25</td></tr> | ||
| + | <tr><td>EcoRI</td><td>Red</td><td>19.75</td><td>.25</td></tr> | ||
| + | <tr><td>EcoRI/SpeI</td><td>Red</td><td>19.5</td><td>.25+.25</td></tr> | ||
| + | <tr><td>XbaI/SpeI</td><td>Tango</td><td>19.5</td><td>.25+.25</td></tr> | ||
| + | <tr><td>EcoRI/PstI</td><td>Red</td><td>19.5</td><td>.25+.25</td></tr> | ||
| + | <tr><td>XbaI/PstI</td><td>Tango</td><td>19.5</td><td>.25+.25</td></tr> | ||
| + | </table><br> | ||
| + | Make up Master Mixes as follows:<br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Red MM</td><td>per tube(µL)</td><td>Total*(µL)</td></tr></b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0</td><td>15.75</td><td>86.675</td></tr> | ||
| + | <tr><td>Red Buffer (10x)</td><td>2</td><td>11</td></tr> | ||
| + | <tr><td>pDNA**</td><td>2</td><td>11</td></tr> | ||
| + | </table><br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Tango MM</td><td>per tube(µL)</td><td>Total*(µL)</td></tr></b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0</td><td>15.75</td><td>86.675</td></tr> | ||
| + | <tr><td>Tango Buffer (10x)</td><td>2</td><td>11</td></tr> | ||
| + | <tr><td>pDNA**</td><td>2</td><td>11</td></tr> | ||
| + | </table> | ||
| + | *Volume per reaction multiplied by 5.5<br> | ||
| + | **Unknown concentration of pDNA<br><br> | ||
| + | Incubated for 70min at 37<sup>o</sup>C (Start-1:05pm; End-2:15pm)<br> | ||
| + | Added 3.3µL of 6x loading dye to each reaction mixture and loaded 10µL onto a 1% agarose gel (in TAE)<br> | ||
| + | Added 1µL of 6x loading dye to 2µL of gene ruler 1kb ladder<br> | ||
| + | Load order as follows:<br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Lane</td><td>Sample</td><td>Volume Loaded (µL)</td></tr></b> | ||
| + | <tr><td>1</td><td>pSB-CEYFP/PstI</td><td>10</td></tr> | ||
| + | <tr><td>2</td><td>pSB-CEYFP/EcoRI</td><td>10</td></tr> | ||
| + | <tr><td>3</td><td>pSB-CEYFP/EcoRI/PstI</td><td>10</td></tr> | ||
| + | <tr><td>4</td><td>pSB-CEYFP/EcoRI/SpeI</td><td>10</td></tr> | ||
| + | <tr><td>5</td><td>pSB-CEYFP/XbaI/PstI</td><td>10</td></tr> | ||
| + | <tr><td>6</td><td>pSB-CEYFP/XbaI</td><td>10</td></tr> | ||
| + | <tr><td>7</td><td>pSB-CEYFP/SpeI</td><td>10</td></tr> | ||
| + | <tr><td>8</td><td>pSB-CEYFP/XbaI/SpeI</td><td>10</td></tr> | ||
| + | <tr><td>9</td><td>pSB-CEYFP/Red Master Mix Control</td><td>10</td></tr> | ||
| + | <tr><td>10</td><td>pSB-CEYFP/Tango Master Mix Control</td><td>10</td></tr> | ||
| + | <tr><td>11</td><td>pSB-CEYFP/MilliQ H<sub>2</sub>0 Control</td><td>10</td></tr> | ||
| + | <tr><td>12</td><td>Ladder</td><td>4</td></tr> | ||
| + | </table><br> | ||
| + | Ran gel at 100V for 1 hour<br><br> | ||
| + | <b>Results:</b><br> | ||
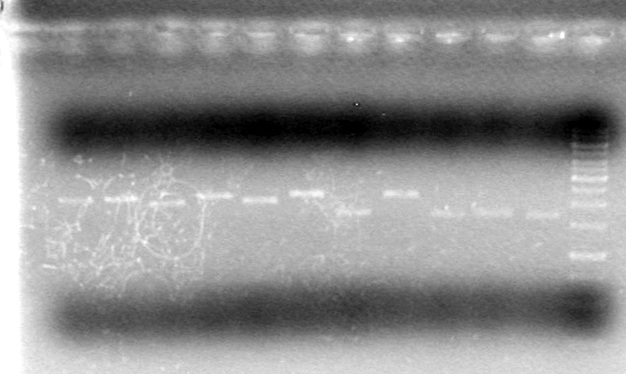
| + | [[image:100505JV-EnzymeTest1Cropped.jpg|200px|none]] | ||
| + | This gel shows that SpeI does not cut on its own, and does not cut when combined with other enzymes<br> | ||
| + | <b>Conclusion:</b> Test other source of SpeI to see if it has any activity.<br><br> | ||
| + | |||
| + | <b><font size=+2>May 6/2010</font>(in the lab:KG, AS)</b><br> | ||
| + | <b>Objective:</b> To check if the old SpeI enzyme (exp date: March 2011) will cleave plasmid DNA, since we believe the newer SpeI enzyme (exp date: 2012) does not.<br> | ||
| + | <b>Method:</b><br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Red Master Mix</b></td><td>per tube (µL)</td><td>Total Volume*</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>15.75</td><td>63</td></tr> | ||
| + | <tr><td>Red Buffer (10x)</td><td>2</td><td>8</td></tr> | ||
| + | <tr><td>pDNA**</td><td>2</td><td>8</td></tr> | ||
| + | </table> | ||
| + | *Volume per tube multiplied by 4<br> | ||
| + | **Used pSB NEYFP pDNA from cell E5 in plasmid box<br> | ||
| + | Enzymes that will use Red Master Mix are: EcoRI+SpeI (old), EcoRI+SpeI (new)<br> | ||
| + | Add 0.25µL of each enzyme to 19.5µL of master mix<br><br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Tango Master Mix</b></td><td>per tube (µL)</td><td>Total Volume*</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>15.75</td><td>94.5</td></tr> | ||
| + | <tr><td>Tango Buffer (10x)</td><td>2</td><td>12</td></tr> | ||
| + | <tr><td>pDNA**</td><td>2</td><td>12</td></tr> | ||
| + | </table> | ||
| + | *Volume per tube multiplied by 6<br> | ||
| + | **Used pSB NEYFP pDNA from cell E5 in plasmid box<br> | ||
| + | Enzymes that will use Tango Master Mix are: SpeI (old), SpeI (new), XbaI+SpeI (old), XbaI+SpeI (new)<br> | ||
| + | Add 0.25µL of each enzyme to 19.5µL of master mix<br><br> | ||
| + | Incubated all reactions at 37<sup>o</sup>C for 1h (Start-8:30pm; End-9:30pm)<br> | ||
| + | Will not be able to run on agarose gel tonight, will label them so JV can run them in the morning<br> | ||
| + | <u>Tube Names:</u><br> | ||
| + | Master Mix 1 Control (Red Buffer)<br> | ||
| + | Master Mix 2 Control (Tango Buffer)<br> | ||
| + | E+S(N); EcoRI + SpeI(N)<br> | ||
| + | E+S(O); EcoRI + SpeI(O)<br> | ||
| + | X+S(N); XbaI + SpeI(N)<br> | ||
| + | X+S(O); XbaI + SpeI(O)<br> | ||
| + | S(N); SpeI(N)<br> | ||
| + | S(O); SpeI(O)<br><br> | ||
| + | Placed in -20<sup>o</sup>C freezer of later analysis by agarose electrophoresis<br><br> | ||
| + | |||
| + | |||
| + | <b><font size=+2>May 10/2010</font>(in the lab:JV)</b><br> | ||
| + | <b>Objective:</b> To analyze the restriction test done by KG and AS on May 6/2010 by agarose electrophoresis<br><br> | ||
| + | <b>Method:</b><br> | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Lane</b></td><td>Sample</td><td>Quantity Loaded (µL)</td></tr> | ||
| + | <tr><td>1</td><td>MM1 Control</td><td>10</td></tr> | ||
| + | <tr><td>2</td><td>MM2 Control</td><td>10</td></tr> | ||
| + | <tr><td>3</td><td>EcoRI+SpeI(N)</td><td>10</td></tr> | ||
| + | <tr><td>4</td><td>EcoRI+SpeI(O)</td><td>10</td></tr> | ||
| + | <tr><td>5</td><td>SpeI(N)</td><td>10</td></tr> | ||
| + | <tr><td>6</td><td>SpeI(O)</td><td>10</td></tr> | ||
| + | <tr><td>7</td><td>XbaI+SpeI(N)</td><td>10</td></tr> | ||
| + | <tr><td>8</td><td>XbaI+SpeI(O)</td><td>10</td></tr> | ||
| + | <tr><td>9</td><td>1kb Ladder</td><td>5</td></tr> | ||
| + | </table> | ||
| + | Run gel for 60min at 100V<br><br> | ||
| + | <b>Results:</b><br> | ||
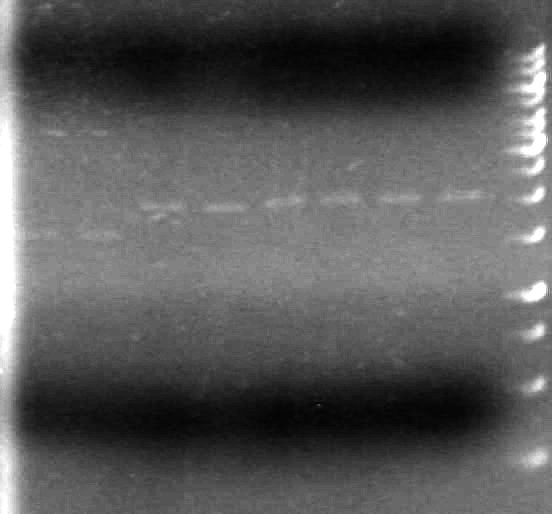
| + | [[image:100510JRV-EnzymeTest1Cropped.jpg|200px|none]]<br> | ||
| + | It appears as though both SpeI enzymes are working properly here. We will utilize the newer batch of SpeI (expires 2012) from this point forward.<br><br> | ||
Revision as of 01:56, 21 May 2010
April 13/2010 (In the Lab: JV, AS)
Objective: Test Restriction Endonucleases for Activity
Relevant Information:
Endonucleases available
| Endonuclease | Optimal Buffer** | Other Buffers |
| EcoRV | None | 2xT(100%); O,G(50-100%) |
| EcoRI | Red | O(100%);R(100%)*;2xT(100%) |
| BcuI/SpeI | Tango | B(50-100%);G(50-100%) |
| XbaI | Tango | B,G,2xT(50-100%) |
| PstI | Orange | R(100%); B,G,T,2xT(50-100%) |
| DpnI | Tango | B,G(100%): O,R,2xT(50-100%) |
- Star Activity
- Optimal Buffer from Fermentas
- Optimal Buffer from Fermentas
Use pUC19 plasmid as test, it has cut sites for EcoRI, PstI, XbaI (unsure about BcuI/SpeI, DpnI but will try anyways), and none for EcoRV
Red Buffer: EcoRI, PstI, Control (No Enzyme)
Tango Buffer: BcuI/SpeI, XbaI, DpnI, Control (No Enzyme>
Methods:
Set up Master Mixes:
| Red MM | per tube (µL) | Total (µL) |
| MilliQ H20 | 13.75 | 55 |
| Red Buffer (10x) | 2 | 7 |
| pUC19 (10pg/µL) | 2 | 7 |
| Total | 19.75 | 69 |
| Tango MM | per tube (µL) | Total (µL) |
| MilliQ H20 | 13.75 | 55 |
| Tango Buffer (10x) | 2 | 7 |
| pUC19 (10pg/µL) | 2 | 7 |
| Total | 19.75 | 69 |
To each tube, add 19.75µL of master mix and 0.25µL of enzyme
Incubated reaction mixes at 37oC (Start:7:00pm; End:7:45pm)
Add 3.3µL of 6x loading dye to each reaction mixture and load 10µL final volume onto a 1% agarose (in TAE) gel.
Add 1µL of 6x loading dye to 1µL of GeneRuler 1kb ladder (at 0.5µg/µL)
Gel loading order as follows:
| Lane | Sample |
| 1 | 1kb Ladder |
| 2 | Tango Control |
| 3 | DpnI (Tango) |
| 4 | BcuI/SpeI (Tango) |
| 5 | XbaI (Tango) |
| 6 | EcoRI (Red) |
| 7 | PstI (Red) |
| 8 | Red Control |
| 9 | Empty |
| 10 | Empty |
Ran gel at 100V for 1 hour
Results: pUC19 plasmid DNA not present at a high enough concentration to visualize by ethidium bromide staining (1kb ladder did stain).
Conclusion: Will have to re-run experiment with DNA that is present at high enough concentrations to visualize by ethidium bromide staining
<a name="may"></a>
May 5/2010(in the lab: JV)
Objective: Test Restriction Endonucleases for activity (take 2)
Relevant Information:
Plasmid DNA used here will be "ES-pSB-CEYFP" from last year's plasmid stocks
Prefix Enzymes are: EcoRI and XbaI
Suffix Enyzmes are: SpeI and PstI
(JV worked out in lab notebook which buffers would be best for each prefix/suffix enzyme combination)
Reactions will be assembled as follows:
| Enzyme</td> | Buffer</td> | Volume MM(µL)</td> | Volume Enzyme(µL)</td></tr> |
| PstI | Red | 19.75 | .25 |
| XbaI | Tango | 19.75 | .25 |
| SpeI | Tango | 19.75 | .25 |
| EcoRI | Red | 19.75 | .25 |
| EcoRI/SpeI | Red | 19.5 | .25+.25 |
| XbaI/SpeI | Tango | 19.5 | .25+.25 |
| EcoRI/PstI | Red | 19.5 | .25+.25 |
| XbaI/PstI | Tango | 19.5 | .25+.25 |
Make up Master Mixes as follows:
| Red MM</td> | per tube(µL)</td> | Total*(µL)</td></tr> |
| MilliQ H20 | 15.75 | 86.675 |
| Red Buffer (10x) | 2 | 11 |
| pDNA** | 2 | 11 |
| Tango MM</td> | per tube(µL)</td> | Total*(µL)</td></tr> |
| MilliQ H20 | 15.75 | 86.675 |
| Tango Buffer (10x) | 2 | 11 |
| pDNA** | 2 | 11 |
- Volume per reaction multiplied by 5.5
- Unknown concentration of pDNA
- Unknown concentration of pDNA
Incubated for 70min at 37oC (Start-1:05pm; End-2:15pm)
Added 3.3µL of 6x loading dye to each reaction mixture and loaded 10µL onto a 1% agarose gel (in TAE)
Added 1µL of 6x loading dye to 2µL of gene ruler 1kb ladder
Load order as follows:
| Lane</td> | Sample</td> | Volume Loaded (µL)</td></tr> |
| 1 | pSB-CEYFP/PstI | 10 |
| 2 | pSB-CEYFP/EcoRI | 10 |
| 3 | pSB-CEYFP/EcoRI/PstI | 10 |
| 4 | pSB-CEYFP/EcoRI/SpeI | 10 |
| 5 | pSB-CEYFP/XbaI/PstI | 10 |
| 6 | pSB-CEYFP/XbaI | 10 |
| 7 | pSB-CEYFP/SpeI | 10 |
| 8 | pSB-CEYFP/XbaI/SpeI | 10 |
| 9 | pSB-CEYFP/Red Master Mix Control | 10 |
| 10 | pSB-CEYFP/Tango Master Mix Control | 10 |
| 11 | pSB-CEYFP/MilliQ H20 Control | 10 |
| 12 | Ladder | 4 |
Ran gel at 100V for 1 hour
Results:
This gel shows that SpeI does not cut on its own, and does not cut when combined with other enzymes
Conclusion: Test other source of SpeI to see if it has any activity.
May 6/2010(in the lab:KG, AS)
Objective: To check if the old SpeI enzyme (exp date: March 2011) will cleave plasmid DNA, since we believe the newer SpeI enzyme (exp date: 2012) does not.
Method:
| Red Master Mix | per tube (µL) | Total Volume* |
| MilliQ H20 Water | 15.75 | 63 |
| Red Buffer (10x) | 2 | 8 |
| pDNA** | 2 | 8 |
- Volume per tube multiplied by 4
- Used pSB NEYFP pDNA from cell E5 in plasmid box
- Used pSB NEYFP pDNA from cell E5 in plasmid box
Enzymes that will use Red Master Mix are: EcoRI+SpeI (old), EcoRI+SpeI (new)
Add 0.25µL of each enzyme to 19.5µL of master mix
| Tango Master Mix | per tube (µL) | Total Volume* |
| MilliQ H20 Water | 15.75 | 94.5 |
| Tango Buffer (10x) | 2 | 12 |
| pDNA** | 2 | 12 |
- Volume per tube multiplied by 6
- Used pSB NEYFP pDNA from cell E5 in plasmid box
- Used pSB NEYFP pDNA from cell E5 in plasmid box
Enzymes that will use Tango Master Mix are: SpeI (old), SpeI (new), XbaI+SpeI (old), XbaI+SpeI (new)
Add 0.25µL of each enzyme to 19.5µL of master mix
Incubated all reactions at 37oC for 1h (Start-8:30pm; End-9:30pm)
Will not be able to run on agarose gel tonight, will label them so JV can run them in the morning
Tube Names:
Master Mix 1 Control (Red Buffer)
Master Mix 2 Control (Tango Buffer)
E+S(N); EcoRI + SpeI(N)
E+S(O); EcoRI + SpeI(O)
X+S(N); XbaI + SpeI(N)
X+S(O); XbaI + SpeI(O)
S(N); SpeI(N)
S(O); SpeI(O)
Placed in -20oC freezer of later analysis by agarose electrophoresis
May 10/2010(in the lab:JV)
Objective: To analyze the restriction test done by KG and AS on May 6/2010 by agarose electrophoresis
Method:
| Lane | Sample | Quantity Loaded (µL) |
| 1 | MM1 Control | 10 |
| 2 | MM2 Control | 10 |
| 3 | EcoRI+SpeI(N) | 10 |
| 4 | EcoRI+SpeI(O) | 10 |
| 5 | SpeI(N) | 10 |
| 6 | SpeI(O) | 10 |
| 7 | XbaI+SpeI(N) | 10 |
| 8 | XbaI+SpeI(O) | 10 |
| 9 | 1kb Ladder | 5 |
Run gel for 60min at 100V
Results:
It appears as though both SpeI enzymes are working properly here. We will utilize the newer batch of SpeI (expires 2012) from this point forward.
 "
"