Apoptosis Notebook
Contents
8-02-2010
8-03-2010
Some test text in bold
We created following tests:
- test4
- test5
8-04-2010
Example of a table
| header 1
| header 2
| header 3
|
| row 1, cell 1
| row 1, cell 2
| row 1, cell 3
|
| row 2, cell 1
| row 2, cell 2
| row 2, cell 3
|
8-05-2010
this too is a table:
| H2Oddes
| 10,3 µl
|
| RE10 + Buffer H
| 2,0 µl
|
| acetylated BSA
| 0,2 µl
|
| DNA
| 6,0 µl
|
table with 3 cells
| apple | banana | peaches
|
| green | yellow | red
|
8-06-2010
text
8-07-2010
text
8-08-2010
test
test
8-09-2010
text Knallroter Text
farbnummern für farbige schrift: http://html.nicole-wellinger.ch/hilfen/farbenverzeichnis.html
test grüner text
8-10-2010
Transforming competent cells
- eGFP Biobrick: BBa_I714891 SDY_eGFP (Kanamycin)
- TEV recogn N Degron SF3 = pDS7 (Ampicillin)
- TEV p14 recogn = 190-6 (Ampicillin)
-> Protocol: (3 Transformation)
- We added 2 µl DNA
- We plated out 200 µl
Plasmid Isolation
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol:(4 Plasmid extraction from cells)
- Prepared overnight culture, measured concentration of DNA
-> Poor results -> thrown away
8-11-2010
New Plasmid Extraction
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol: (4 Plasmid extraction from cells)
- Plasmid concentration: 143ng/µl
Prepared overnight culture of eGFP BBa_I714891
- 3 ml LB-Media + 4 µl Kanamycin
- Inoculated iangeimpft) with 1 colony of BBa_I714891 -> 37°C
Prepared overnight culture of 190-6 and pDS7 and eGFP (BBa_I714891) in falcons
- for 190-6 and pDS7: 10µl Ampicillin + 10 ml LB-Media + colony of plate
- for eGFP: 13,3 µl Kanamycin + 10 ml LB-Media + 1 colony of plate
Restriction digestion of CMV-Promoter BBa_J52034 with EcoRI and PstI
| H2Oddest, sterile
| 10,3 µl
|
| RE10 + Buffer H
| 2,0 µl
|
| acetylated BSA (18ng/µl)
| 0,2 µl
|
| DNA (0,143µg/µl)
| 6,0 µl
|
-> mixed
- plus: EcoRI (10µg/µl): 0,5 µl resp. PstI (10µg/µl): 0,5 µl
- incubated at room temperature from 12:10 to 15:00, 1 hour at 37°C, 2 hours at 60°C
- frozen at -20°C
Prepare new/fresh overnight culture of CMV-Promoter Biobrick: BBa_J52034
- 1 ml of "old" culture + 3 ml LB-Media + 4 µl Kanamycin -> 37°C
8-12-2010
Plasmid Extraction of pDS7, eGFP, 190-6
-> Protocol: (4 Plasmid extraktion from cells)
- pDS7 (458ng/µl), eGFP (55ng/µl), 190-6 (193ng/µl)
Restriction digest of pDS7, eGFP, 190-6
- with EcoRI and PstI in buffer H (for testing DNA is correct)
-> Protocol: (5 Restriction digest)
- 10µg DNA: pDS7 (2µl), eGFP (15µl), 190-6 (10µl)
Plate colonies for plasmid extrction
- CMV (Kanamycin), eGFP (Kanamycin), pDS7 (Ampicillin), 190-6 (Ampicillin))
- PhiC31o plated on Ampicillin-Agar, stored at 37°C
50% Glycerol made
- for PhiC31o glycerol stock (produced later)
8-13-2010
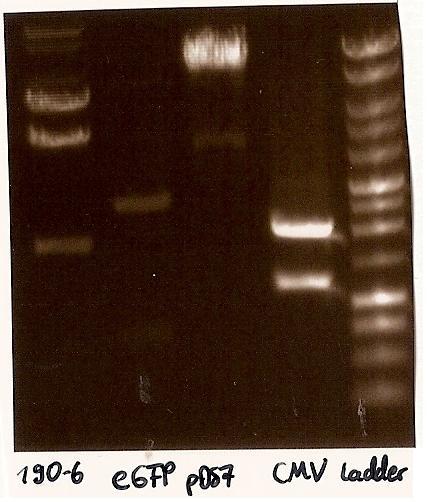 Gelfoto from the EcoR1 and Pst1 Restrictiondigest of 190-6, eGFP, pDS7 and CMV Inoculate CMV into LB medium with amicillin
- CMV (BBa_J52034) from 10.8.2010 inoculated into LB medium with ampicillin, as falsly inoculated in Kanamycin
Agarosegelelectrophoresis with digestions
->Protocol (11 Agarose gel electrophoresis)
- Agarosegelelectrophoresis with the digestions (CMV, eGFP, pDS7, 190-6), 125V for 30 minutes and then for 20 minutes;
- expected DNA bands: 190-6 (4840bp, 1903bp), pDS7 (8027bp, 6bp), CMV (654 bp (Insert), 2079bp (Plasmid)), eGFP (720bp (Insert), 2750bp (Plasmid))
- Correct DNA bands for 190-6 (~4800bp, ~1900bp, ~6700bp (undigested plasmid)) and eGFP (~2000bp (Plasmid), ~750 bp (Insert)); CMV probably not digested (two bands; one probably normal, one supercoiled) and pDS7 not clear
Restriction digest from CMV and pDS7
-> Protocol (5 Restriction digest)
- Restriction digest from CMV (EcoR1, Pst1; 6µl DNA, buffer H) and pDS7 (EcoR1, Spe1; 2µl DNA, buffer B)
Agarosegelectrophoresis with digestions
->Protocol (11 Agarose gel electrophoresis)
- Agarosegelelectorphoresis for 30 minutes, 150V
- Expected DNA bands: CMV see above, pDS7 (3647bp, 3369bp, 1011bp, 6bp)
- false DNA bands CMV (~1200 bp, ~2000 bp) and pDS7 (~8000bp two bands, ~1100 bp); required to isolate a new colony for these two Plasmidextractions
Plated CMV on Ampicllin-Agar
- Plated the colony from CMV (BBa_J52034) for Plasmidextraction (Ampicillin), as falsly plated on Kanamycin
8-14-2010
weekend
8-15-2010
weekend
8-16-2010
Planting colonies
- transfer 1 ml PhiC31o culture to new LB medium + Amp, 37°C
- pick up CMV and pDS7 colonies from plates and transfer to LB medium+Amp, 37°C
Plasmid Extraction of PhiC31o
->Protocol (4 Plasmid extraktion from cells)
- plasmid extraction of PhiC310
->27,5ng/µl DNA and second plasmid extraction of PhiC310 (i. o. to get more DNA); first eluation-step with first eluation-extraction
-> 60ng/µl DNA
Restriction digest
->Protocol (5 Restriction digest)
- restriction digest of PhiC310 with EcoR1 and Spe1
| H2Oddest, sterile
| 0 µl
|
| Buffer B
| 2,0 µl
|
| BSA (1:10)
| 2 µl
|
| DNA (0,06µg/µl)
| 15,0 µl
|
| EcoR1
| 0,5 µl
|
| Spe1
| 0,5µl
|
restriction digest in the thermo cycler (program "Verdau", see protocol)
Handling primers after arrival (1,2,3,4,5,6,11,12)
->Protocol (9 Handling primers)
PCR preparations
- 10mM dNTP mix made from 100 mM dATP, dGTP, dCTP, dTTP by taking 100µl of each and adding 600µl H 2 O
PCR 1 and 6
- PCR of the tet inducible CMV minimal promotor out of prevTRE (=PCR 1 with Primer 1 and 2) and SV40PA out of pcDNA3 (=PCR 6 with Primer 11 and 12)
->Protocol (10 PCR with Pfu)
Mixture:
|
| pTRERev (0,15µg/µl)
| pcDNA3 (0,6 µg/µl)
|
| Primer
| 2*2,5µl (P1+P2)
| " (P11+P12)
|
| 300ng template
| 0,5µl
| 2µl
|
| 10x Buffer Pfu
| 5µl
| "
|
| dNTP Mix
| 1µl
| "
|
| Pfu Polymerase (3u/µl)
| 0,5µl
| "
|
| H2O
| 40,5µl
| 39µl
|
| summ
| 52,5µl
| 52,5µl
|
Programme:
| Denaturation
| 95°C
| 2min
|
| 30 times:
| Denaturation
| 95°C
| 1min
|
|
| Annealing
| 45°C
| 30sec
|
|
| Extension
| 73°C
| 2min
|
| Final Extension
| 73°C
| 5min
|
| Soak (end)
| 12°C
| infinite
|
Glycerolstock of PhiC31o
- Glycerolstock of the colony of PhiC31o for the plasmidextraction
| bacterial culture
| 800µl
|
| Glycerol (50%)
| 500µl
|
8-17-2010
 Agarose gel electrophoresis of PCR6 which shows that PCR6 is about 200bp Plate CMV and pDS7 colonies on Ampicillin-Agar
- colonies for plasmidextraction of CMV and pDS7 plated on Ampicillinplates
Plasmid Extraction of CMV and pDS7
- plasmidextraction of CMV (2,5ng/µl) and pDS7 (10ng/µl) the A260/A280 value was 1.333, which means that it was 90% Protein and only 10% DNA (should be 1,8); new plasmidextraction needed
new overnight cultures of CMV and pDS7 for a new plasmidextraction made
Agarose gel electrophoresis
-> Protocol (11 Agarose gel electrophoresis)
- Agarose gel electrophoresis of the restriction digest of PhiC31o and PCR 1 and 6
- the right bands found for PhiC31o (~2900,~2400,~250)
- the right band found for PCR1 (~450)
- no band found for PCR6; new electrophoresis needed with more DNA loaded

|

|
|
Agarose gel electrophoresis of (from left to right) PhiC31o, PCR1 and PCR6
|
Agarose gel electrophoresis of (from left to right) PhiC31o, PCR1 and PCR6 which shows that PCR1 is between 250 and 500 bp
|
- new agarose gel electrophoresis from PCR6 with 5µl DNA instead of 3µl (image not yet shown)
- the right band found for PCR6 (~200)
New overnight cultures of CMV and pDS7
- the overnight colonies didn't grow; new colonies (CMV and pDS7) picked from plate and inoculated in LB Ampicillin
PCR purification of PCR 1 and 6
-> Protocol (12 Gel extraction or PCR Clean up)
- DNA concentration of the PCR 1 and 6 products measured: PCR1: 410ng/µl (A260/A280=1.253) PCR6: 568ng/µl (A260/A280=1.275)
- PCR Purification with Promega Kit
-> PCR1: 230ng/µl (A260/A280=1.769)
-> PCR6: 37.5ng/µl (A260/A280=1.667)
8-18-2010
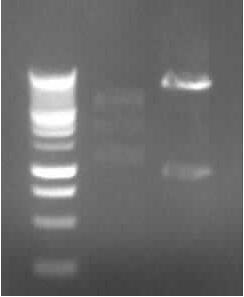 Agarose gel electrophoresis of (from left to right) CMV and pDS7 showing the right bands for pDS7 Plasmid Extraction of CMV and pDS7
-> Protocol (4 Plasmid extraction from cells)
- Plasmid extraction of CMV (97.5ng/µl; A260/A280=1.857) and pDS7 (212ng/µl; A260/A280=1.848)
Restriction digestion
-> Protocol (5 Restriction digestion)
- Restriction digestion of CMV (EcoR1 + Pst1; 10µl DNA, buffer H) and pDS7 (EcoR1 + Spe1; 5µl DNA, buffer B)
-> expected DNA bands: CMV: 2079bp (plasmid) + 654bp (Insert); pDS7: 7022bp + 1011bp
Agarose Gel electrophoresis of digested CMV and pDS7
-> Protocol (11 Agarorse gel electrophoresis)
-> right DNA bands for pDS7 (~7000bp, ~1000bp)
-> false DNA bands for CMV
- Starting PCR 2a and 2b (replication and mutagenesis of pDS7): 3 µl DNA and 50°C Annealing Temperatur (other same as 8-16-2010)
8-19-2010
Agarose gel electrophoresis of PCR 2a and 2b
-> Protocol (11 Agarose gel electrophoresis)
(150V, 30min)
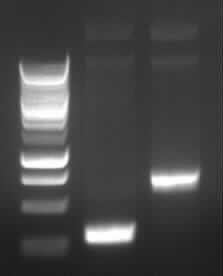
|
| Agarose gel electrophoresis of (from left to right) PCR2a and PCR2b
|
-> the right bands for PCR2a (~300bp) and PCR2b (~700bp)
- New agarose gel electrophoresis with all of the PCR product for gel extraction (150V, 30min)
Gel extraction of the DNA from PCR2a and PCR2b
-> Protocol (12 Gel extraction or PCR Clean up)
- DNA concentration measured; problem with nanodrop as too low concentration; lyophille used to reduce volume
- DNA concentration measured again: PCR2a: 70ng/µl A260/A280=1.647; PCR2b: 45ng/µl A260/A280=1.5
PCR 3 (joining PCR of 2a and 2b)
- PCR3 (the joining PCR of PCR2a and 2b; Joining of the TEVrecogn-N-Degron-SF3 part) done: 1.3 µl of PCR2a and 4.7 µl of PCR2b makes 300ng of a 1:1 solution of both to be joined DNA parts. Annealing temperature: 50°C
-> Protocol (10 PCR with Pfu)
8-20-2010
Agarose gel electrophoresis of PCR3
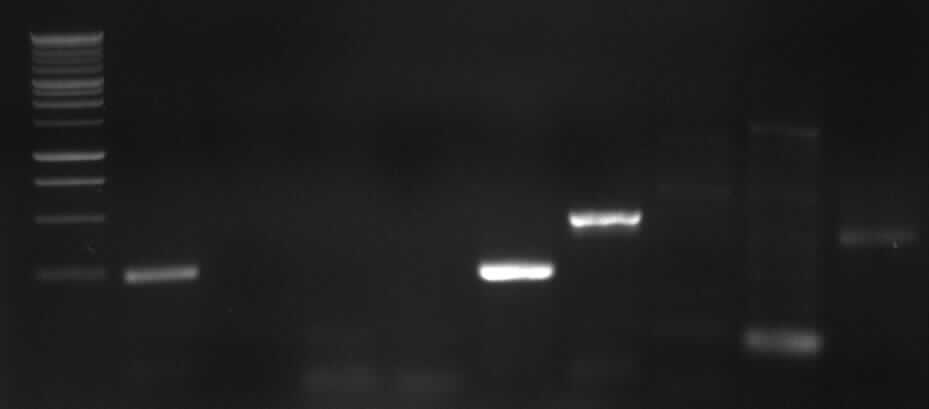
left column: marker; most right column: PCR3
-> Protocol: 11 Agarose gel electrophoresis (150V, 30min)
-> expected band: ~1000bp
-> false band: ~500bp
- probable reason: mini photometre was influenced by gel extraction chemicals, therefore it measured false DNA concentrations and false template masses were calculated
-> New 2a and 2b PCR
New PCR (2a and 2b)
-> Protocol: 10 PCR with Pfu
(see 8-18-2010, but 35,5µl water)
8-21-2010
weekend
8-22-2010
weekend
8-23-2010
Agarose gel electrophoresis of PCR 2b
-> Protocol: 11 Agarose gel electrophoresis
- expected band: 700bp
-> no band shown on gel -> new PCR 2b
PCR 2b
- start PCR 2b with PCR 2b from 8-13-10 as template ( 1:20 and 1:100 diluted; 1µl)
-> Protocol: 10 PCR with Pfu
- annealing temperature: 50°C; amount of water: 37,5µl
Agarose gel electrophoresis of PCR 2b 1:20 and 1:100
-> Protocol: 11 Agarose gel electrophoresis
- expected bands: each ~ 700bp
- false bands: ~ 200bp
-> new PCR with 2ng, 5ng, 10ng template pDS7
PCR 2b with 2ng, 5ng, 10ng template pDS7
- pDS7 1:100 diluted(-> 2,1 ng/µl)
Mixture:
|
| 2ng
| 5ng
| 10ng
|
| Primer
| 2*2,5µl (P5+P6)
| 2*2,5µl (P5+P6)
| 2*2,5µl (P5+P6)
|
| 10x Buffer Pfu
| 5µl
| 5µl
| 5µl
|
| dNTP Mix
| 1µl
| 1µl
| 1µl
|
| template
| pDS7 (dil.)
| 1µl
| 2,5µl
| 5µl
|
| Pfu Polymerase (3u/µl)
| 0,5µl
| 0,5µl
| 0,5µl
|
| DMSO
| 1,25µl
| 1,25µl
| 1,25µl
|
| H2O
| 33,25µl
| 30,25µl
| 25,25µl
|
| sum
|
|
|
|
-> Protocol: 10 PCR with Pfu
PCR 2a gel extraction
- Quaigen kit (QuaiexII)
-> Protocol: 14 QIAEX II gel extraction
Start 3 CMV overnight cultures
8-24-2010
agarose gel electrophoresis of PCR 2b
-> Protocol: 11 Agarose gel electrophoresis
 Agarose gel electrophoresis of (from left to right) CMV (2ng (cut out), 10ng, 5ng template) showing the right bands for 2ng, 5ng template - expected bands: right bands with 2ng and 5ng template (~700bp), no band with 10ng template
CMV plasmid extraction
-> Protocol: 4 Plasmid extraction from cells
Plasmid extractionof 3 different overnight cultures.
- results:
- 52,5 ng/µl A260/A280= 1.312
- 133 ng/µl A260/A280= 1.710
- 80 ng/µl A260/A280= 1.600
CMV restriction digestion
-> Protocol: 5 Restriction digestion
- CMV restriction digest: EcoRI, PstI, buffer H
- 19µl, H2O : 0µl
- 6µl, H2O : 9.5µl
- 12.5µl, H2O : 3µl
PCR 2b gel extraction
- PCR2b was gel extracted (with Qiagen gel extraction kit), 17.5 ng/µl a260/A280= 1.750
-> Protocol: 14 QIAEX II gel extraction
PCR 3 (fusion of 2a and 2b)
- PCR3: conducted again at 52°C annealing temperature
- 10.5 ng (from PCR2b) 0.6µl
- 4.5 ng (from PCR2a) 0.9µl (1:10 diluted)
| PCR2a
| 0.9 µl
|
| PCR2b
| 0.6 µl
|
| primer3
| 2.5 µl
|
| primer6
| 2.5 µl
|
| dNTPs
| 1 µl
|
| Pfu
| 0.5 µl
|
| 10xbuffer
| 5 µl
|
| H2O
| 37 µl
|
-> Protocol: 10 PCR with Pfu
agarose gel electrophoresis of CMV digestion
- agarose gel electrophoresis (150V, 25 min) of the CMV digestion
-> bands are wrong again ( ~ 1200bp, 2000bp)
8-25-2010
Agarose gel electrophorese of PCR 3
-> Protocol: 11 Agarose gel electrophoresis
- expected band: ~1000bp
- false band: ~400bp
Plasmid extraction of ccdB tet and ccdB strep
Plasmid extraction of pSB1C3 with BBa_P1010
-> Protocol: 4 Plasmid extraction from cells
- results:
| ccdB tet:
| 50ng/µl;
| A260/A280= 1,818
|
Plate ccdB amp, cam, tet
- plate ccdB with ampicilline, chloramphenicol, tetracycline resistence on LB agar with appropiate antibiotic.
Overnight culture of ccdB kan
- Overnight culture of ccdB with kanamycine resistence in LB medium with kanamycine
PCR 7a, 7b, 9, 10
->Protocol: 10 PCR with Pfu
| PCR nr.
| template
| concentration
| dilution
| primer
|
| 7a
| 190-6
| ~200ng/µl
| 1:100
| 13,14
|
| 7b
| 190-6
| ~200ng/µl
| 1:100
| 15,16
|
| 9
| eGFP
| 55ng/µl
| 1:25
| 20,21
|
| 10
| PhiC31o
| 20ng/µl
| 1:10
| 22,23
|
Mixture
| template (~2ng)
| 1µl
|
| Pfu
| 0,5µl
|
| Primer *2
| 2,5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| H2O
| 37,5µl
|
| sum
| 50µl
|
Standard PCR; annealing temperature: 60°C
8-26-2010
Agarose gelelectrophoresis of PCR 7a, 7b, 9, 10
->Protocol: 11 Agarose gel electrophoresis
- 150V, 25min
| PCR nr.
| expected bands
| result
|
| 7a
| 850bp
| no band
|
| 7b
| 402bp
| false band (200bp)
|
| 9
| 808bp
| no band
|
| 10
| 1888bp
| no band
|
Plasmid extraction of ccdB kan
-> Protocol: 4 Plasmid extraction from cells
-result: concentration: 25ng/µl; A260/A280= 2,0
New PCR 7a, 7b, 9, 10
Mixture
| template (~2ng)
| 1µl
|
| Pfu
| 0,5µl
|
| Primer *2
| 2,5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 36,25µl
|
| sum
| 50µl
|
Program:
gradient PCR, 42-69°C annealing temp.
->Protocol: 10 PCR with Pfu
Overnight culture of ccdB amp, tet, cam
Inoculate one colony each in 5ml medium with approptraite antibiotic.
8-27-2010
Agarose gel electrophoresis of PCR 7a, 7b, 9, 10
->Protocol: 11 Agarose gel electrophoresis
150V, 25min, 75mA
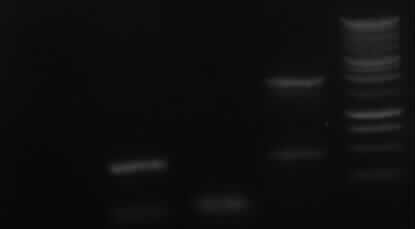
from left to right: 7a, 7b, 9, 10, Marker
| PCR nr.
| expected bands
| result
|
| 7a
| 850bp
| no band
|
| 7b
| 402bp
| right band (~400bp)+ false band (~150bp)
|
| 9
| 808bp
| false band (~200bp)
|
| 10
| 1888bp
| right band (~1900bp)+false band (~500bp)
|
Plasmid extraktion of ccdB amp, tet, cam
->Protocol: 4 Plasmid extraction from cells
results:
| Plasmid
| concentration
| A260/A280
|
| ccdB amp
| 57,5 ng/µl
| 1,917
|
| ccdB cam
| 70,0 ng/µl
| 1,867
|
| ccdB tet
| 50,0 ng/µl
| 1,818
|
New PCR 7a, 9
Mixture:
- 2ng template: see 26-8-10
- 4ng template: see 26-8-10, but 2µl template and 35,25µl water
-> Protocol: 10 PCR with Pfu
Restriction digestion of ccdB amp, kan, cam, tet
-> Protocol: 5 Restriction digestion
- only 90min 37°C incubation
- EcoRI, PstI, Buffer H
| template
| volume
| mass
|
| ccdB amp
| 16µl
| 930ng
|
| ccdB cam
| 14,3µl
| 1µg
|
| ccdB tet
| 16µl
| 800ng
|
| ccdB kan
| 16µl
| 400ng
|
Agarose gelelectrophoresis of PCR 7a, 9, ccdB restriction digestion
150v, 25min, 75mA
-> Protocol: 11 Agarose gel electrophoresis
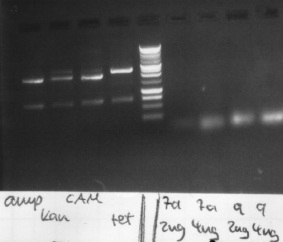
results:
- PCR7a, 9: false band at 200bp
- ccdB: each digestion leads to a right band with ~ 650bp
8-28-2010
weekend
8-29-2010
weekend
8-30-2010
New PCR 7a and 9
Mixture
| template (~4ng)
| 2µl
|
| Pfu
| 0,5µl
|
| Primer *2
| 2,5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 35,25µl
|
| sum
| 50µl
|
-> Protocol: 10 PCR with Pfu
PCR program
PCR 7: Annealing Temperature 60°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time
PCR 9: Annealing Temperature 55°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time
Gel extraction of PCR 7b, 10
->Protocol: 14 QIAEX II gel extraction
results:
| PCR nr.
| concentration
| A260/A280
|
| 7b
| 10 ng/µl
| 2,0
|
| 10
| 17,5 ng/µl
| 1,4
|
Agarose gel electrophoresis of new PCR 7a, 9
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
- results:
- 7a: no band shown
- 9: false band (~200bp)
New PCR 3
-> Protocol: 10 PCR with Pfu
| PCR2a
| 0.9 µl
|
| PCR2b
| 0.6 µl
|
| dNTPs
| 1 µl
|
| Pfu
| 0.5 µl
|
| 10xbuffer
| 5 µl
|
| DMSO
| 1,25µl
|
| H2O
| 36,75 µl
|
| sum
| 45µl
|
- new method: standard PCR without primers (10 cycles, 56°C annealing temp.)
- then add 2,5µl of primer 3 and 6
- 30 cycles standard PCR (54°C annealing temp.)
8-31-2010
 Gel photo of (left to right) PCR4a(2.5ng template), PCR4a(5ng template),PCR4b(2.5ng template), PCR4b(5ng template), PCR3(Pfu), PCR3(Phusion) Agarose gel electrophoresis of PCR 4a, PCR4b, PCR3(Pfu), PCR3(Phusion)
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
- results:
- PCR4a(2.5ng template), PCR4a(5ng template),PCR4b(2.5ng template), PCR4b(5ng template), PCR3(Pfu): no band shown
- PCR3 (Phusion): right band (~1000bp)
New PCR PCR4a, PCR4b, PCR7a, PCR9
-> Protocol: 10 PCR with Pfu
PCR mixture for PCR4a, PCR4b
| template
| 37.25 µl (200ng)
|
| dNTPs
| 1 µl
|
| Pfu
| 0.5 µl
|
| 10xbuffer
| 5 µl
|
| DMSO
| 1,25µl
|
| sum
| 50µl
|
Standard PCR program with annealing temperature PCR4a: 51.1°C, PCR4b: 48.5°C.
-> Protocol: 15 PCR with Phusion
PCR mixture for PCR7a, PCR9
| template
| 4 µl (8ng)
|
| dNTPs
| 1 µl
|
| Phusion
| 0.5 µl
|
| 5xbuffer
| 10 µl
|
| DMSO
| 1,25µl
|
| H2O
| 28.25 µl
|
| sum
| 50µl
|
PCR program: Phu62
| 98°C
| 1 min
|
| 98°C
| 10 sec
|
| 62°C
| 20 sec
|
| 73°C
| 30 sec
|
| return to step 2 for 29 cycles
|
|
| 73°C
| 10 min
|
| 12°C
| forever
|
 Gel photo of (left to right) PCR3, ladder, PCR7a, empty, PCR9 Agarose gel electrophoresis of PCR 3, PCR7a, PCR9 for gel extraction
-> Protocol: 11 Agarose gel electrophoresis
120V, 30min
- results:
- PCR3; right band (~1000bp) and side-product
- PCR7a: no band
- PCR9: right band (~800bp) and side-product
Gel extraction of the DNA from PCR3 and PCR9
-> Protocol (12 Gel extraction or PCR Clean up)
results:
PCR 9: 22,5ng/µl; A260/A280=1,8
PCR 3: 22,5ng/µl; A260/A280=2,25
New PCR 4a, 4b, 7a with DreamTaq
-> Protocol: 16 PCR with DreamTaq
PCR mixture for PCR7a
| template
| 5 µl (10ng)
|
| dNTPs
| 5 µl
|
| DreamTaq
| 0.33 µl
|
| 10xbuffer
| 5 µl
|
| DMSO
| 1,25µl
|
| H2O
| 28.5 µl
|
| sum
| 50µl
|
Primers for PCR 7a: 13,14
Annealing temp: 60°C
PCR mixture for PCR4a,4b
| template
| 36,5 µl (180ng HeLa cDNA)
|
| dNTPs
| 5 µl
|
| DreamTaq
| 0.33 µl
|
| 10xbuffer
| 5 µl
|
| DMSO
| 1,25µl
|
| sum
| 50µl
|
PCR 4a
Primers for PCR 4a: 7,8
Primers for PCR 4b: 9,10
Annealing temp: 50°C
PCR program:
| 95°C
| 1 min
|
| 95°C
| 30 sec
|
| 50/60°C
| 30 sec
|
| 72°C
| 1 min (1kb/min)
|
| return to step 2 for 29 cycles
|
|
| 72°C
| 10 min
|
| 12°C
| forever
|
9-01-2010
Agarose gel electrophoresis of PCR4a, PCR4b, PCR7 (DreamTaq)
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
results: no product
New PCR 4a, 4b with DreamTaq, Pfu, with concentration gradient and touch-down PCR
-> Protocol: 16 PCR with DreamTaq; 10 PCR with Pfu
PCR mixture for DreamTaq
| Concentration
| Low
| Middle
| High
|
| template
| 5 µl (1:1000)
| 31.75µl (1:1000)
| 2µl (1:10)
|
| dNTPs
|
| 5 µl
|
|
| DreamTaq
|
| 0.33 µl
|
|
| 10xbuffer
|
| 5 µl
|
|
| DMSO
|
| 1,25µl
|
|
| H2O
| 26.75 µl
| 0
| 29.75
|
| sum
|
| 50µl
|
|
PCR mixture for Pfu
| Concentration
| Low
| Middle
| High
|
| template
| 5 µl (1:1000)
| 37.25µl (1:1000)
| 2µl (1:10)
|
| dNTPs
|
| 1 µl
|
|
| Pfu
|
| 0.5 µl
|
|
| 10xbuffer
|
| 5 µl
|
|
| DMSO
|
| 1,25µl
|
|
| H2O
| 32.25 µl
| 0
| 35.25 µl
|
| sum
|
| 50µl
|
|
Primers for PCR 4a: 7,8; PCR 4b: 9,10
-> Protocol: Thermal cycler program: Touch down
Agarose gel electrophoresis of PCR4a, PCR4b
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
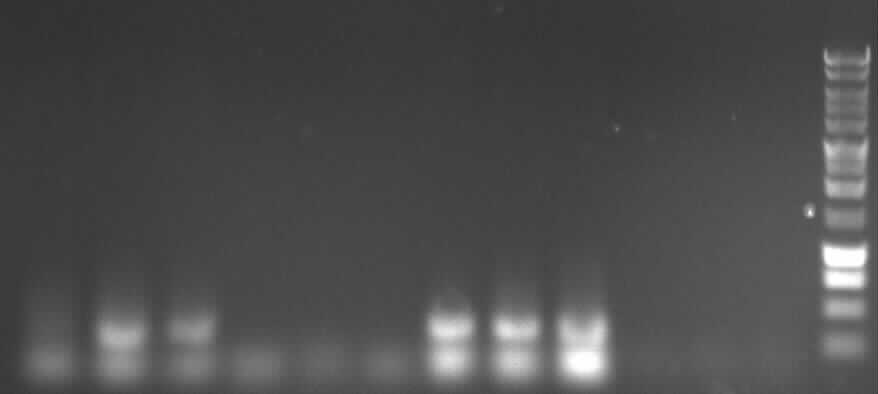
from left to right: 4a: P1, P2, P3, D1, D2, D3; ab: P1, P2, P3, D1, D2, D3
key:
"P"= PCR with Pfu
"D"= PCR with DreamTaq
"1"= low template concentration
"2"= middle template concentration
"3"= high template concentration
expected bands:
- 4a: 330bp -> P2 and P3 show right bands and "primer clouds"(?)
- 4b: 376bp -> P1, P2, P3 show right bands and "primer clouds" (?)
9-02-2010
Agarose gel electrophorese of PCR 4a P2, 4b P2
-> Protocol: 11 Agarose gel electrophoresis
- 120V, 45min, 1,5% Agarose gel

Agarose gel electrophoresis of (from left to right) PCR4aP2, Marker and PCR4bP2
- Cut out bands at ~~ 350bp and extract
PCR Agarose gel extraction
-> Protocol: 14 QIAEX II gel extraction
results:
| PCR nr.
| concentration
| A260/A280
|
| 4aP2
| 12,5 ng/µl
| 1,67
|
| 4bP2
| 72,5 ng/µl
| 1,53
|
New PCR 7a with Pfu and Phusion
template: 190-6, Primer 13,14
Mixture with Pfu
| template (~2ng)
| 1µl
|
| Pfu
| 0,5µl
|
| Primer *2
| 2,5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 36,25µl
|
| sum
| 50µl
|
-> Protocol: 10 PCR with Pfu
PCR mixture with Phusion
| template (~2ng)
| 1,0 µl
|
| dNTPs
| 1 µl
|
| Phusion
| 0.5 µl
|
| 5xbuffer
| 10 µl
|
| DMSO
| 1,25µl
|
| H2O
| 31,25µl
|
| sum
| 50µl
|
-> Protocol: 15 PCR with Phusion
New PCR 4a, 4b with Pfu
-> Protocol: 10 PCR with Pfu
-> Protocol: Thermal cycler program: Touch down
Mixture see 9-1-10, twice 4a and 4b
Agarose gel electrophorese of PCR 4a, 4b, 7a, gel extracted 4a and 4b
-> Protocol: 11 Agarose gel electrophoresis
- 25min, 150V
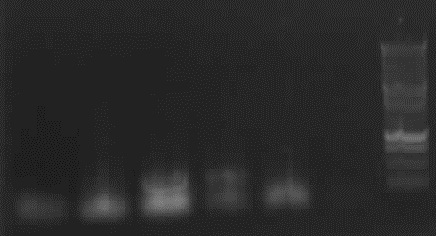
from left to right: 4a*, 4a, 4b*, 4b, 7a Phusion, 7a Pfu, Ladder
-> result: 4b, 4b*: right bands (~330bp)
-remain: false bands/no band
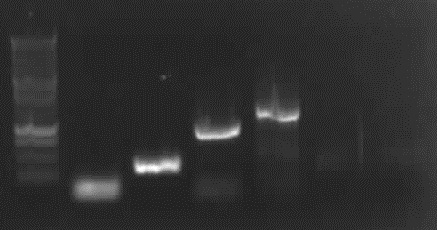
from left to right: ladder, 4 columns pathway, 4a gelextr., 4b gelextr.
-> results: slight right bands for 4a and 4b, no "primer clouds" anymore.
9-03-2010
Overlapping PCR 5 with Pfu and Phusion
template: 4a, 4b Primer 7,10
Mixture with Pfu
| PCR4a (~15ng)
| 1.2µl
|
| PCR4b (~15ng)
| 2µl (1:10)
|
| Pfu
| 0.5µl
|
| Primer *2
| 2.5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 34.05µl
|
| sum
| 50µl
|
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Annealing temperature: 54°C
PCR mixture with Phusion
| PCR4a (~15ng)
| 1.2µl
|
| PCR4b (~15ng)
| 2µl (1:10)
|
| dNTPs
| 1 µl
|
| Phusion
| 0.5 µl
|
| 5xbuffer
| 10 µl
|
| DMSO
| 1,25µl
|
| H2O
| 29.05µl
|
| sum
| 50µl
|
-> Protocol: 15 PCR with Phusion
PCR program: standard PCR program for Phusion, Annealing temperature: 58°C
New PCR7a with Pfu
template: 190-6; Primer: 13,14
Mixture with Pfu
| template (~2ng)
| 1µl
|
| Pfu
| 0.5µl
|
| Primer *2
| 2.5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 36.25µl
|
| sum
| 50µl
|
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Gradient: 54.4°C, 57.8°C, 61.4°C, 65.0°C
-> results:
-7a: only "primer cluods"
-5 Pfu: no defined product (slurred?)
-5 Phusion: "primer clouds"
9-04-2010
weekend
9-05-2010
weekend
9-06-2010
charges for sequencing
| name
| 4a-7
| 4a-8
| 4b-9
| 4b-10
| 3-3
| 3-6
| 6-11
| 6-12
|
| DNA
| 4a; 2.4µl
| 4a; 2.4µl
| 4b; 0.5µl
| 4b; 0.5µl
| 3; 2µl
| 3; 2µl
| 6; 0.5µl
| 6; 0.5µl
|
| primer [1pmol/µl]
| 7; 3.2µl
| 8; 3.2µl
| 9; 3.2µl
| 10; 3.2µl
| 3; 3.2µl
| 6; 3.2µl
| 11; 3.2µl
| 12; 3.2µl
|
| Tris (10mM); pH 8.2
| 1.4µl
| 1.4µl
| 3.3µl
| 3.3µl
| 1.8µl
| 1.8µl
| 3.3µl
| 3.3µl
|
every charge 7µl
New overlapping PCR 5 with Pfu
template: 4a, 4b; 15ng
Primer 7,10
Mixture
| PCR4a (~7ng)
| 0.56µl
|
| PCR4b (~8ng)
| 1.1µl (1:10)
|
| Pfu
| 0.5µl
|
| Primer *2
| 2.5µl *2
|
| 10x buffer
| 5µl
|
| dNTP Mix
| 1µl
|
| DMSO
| 1,25µl
|
| H2O
| 36.6µl
|
| sum
| 50µl
|
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Annealing temperature: 46.1°C,48.5°C, 51.1°C
-> results: "primer clouds"
New PCR 7a with Pfu
new dilution of 190-6; 1:100
| template[2ng/µl]
| 1µl
| 2µl
|
| pfu polymerase
| 0.5µl
| 0.5µl
|
| primer13
| 2.5µl
| 2.5µl
|
| primer14
| 2.5µl
| 2.5µl
|
| buffer
| 5µl
| 5µl
|
| dNTP Mix
| 1µl
| 1µl
|
| DMSO
| 1,25µl
| 1.25µl
|
| H2O
| 36.25µl
| 35.25
|
| sum
| 50µl
| 50µl
|
Agarose gel electrophoresis of 7a
-> results: "primer clouds"
charges for sequencing for PCR 1,7b and 9
| name
| 1-1
| 1-2
| 7b-15
| 7b-16
| 9-20
| 9-21
| 10-22
| 10-23
|
| DNA
| 1[1:10]; 1.74µl
| 1[1:10]; 1.74µl
| 7b; 3µl
| 7b; 3µl
| 9; 1.78µl
| 9; 1.78µl
| 10; 3.8µl
| 10; 3.8µl
|
| primer [1pmol/µl]
| 1; 3.2µl
| 2; 3.2µl
| 15; 3.2µl
| 16; 3.2µl
| 20; 3.2µl
| 21; 3.2µl
| 22; 3.2µl
| 23; 3.2µl
|
| Tris (10mM); pH 8.2
| 2.06µl
| 2.06µl
| 0.8µl
| 0.8µl
| 2.02µl
| 2.02µl
| 0µl
| 0µl
|
every charge 7µl
9-07-2010
Agarose gel electrophorese of PCR gel extractions
150V, 25min
-> Protocol: 11 Agarose gel electrophoresis
-result:
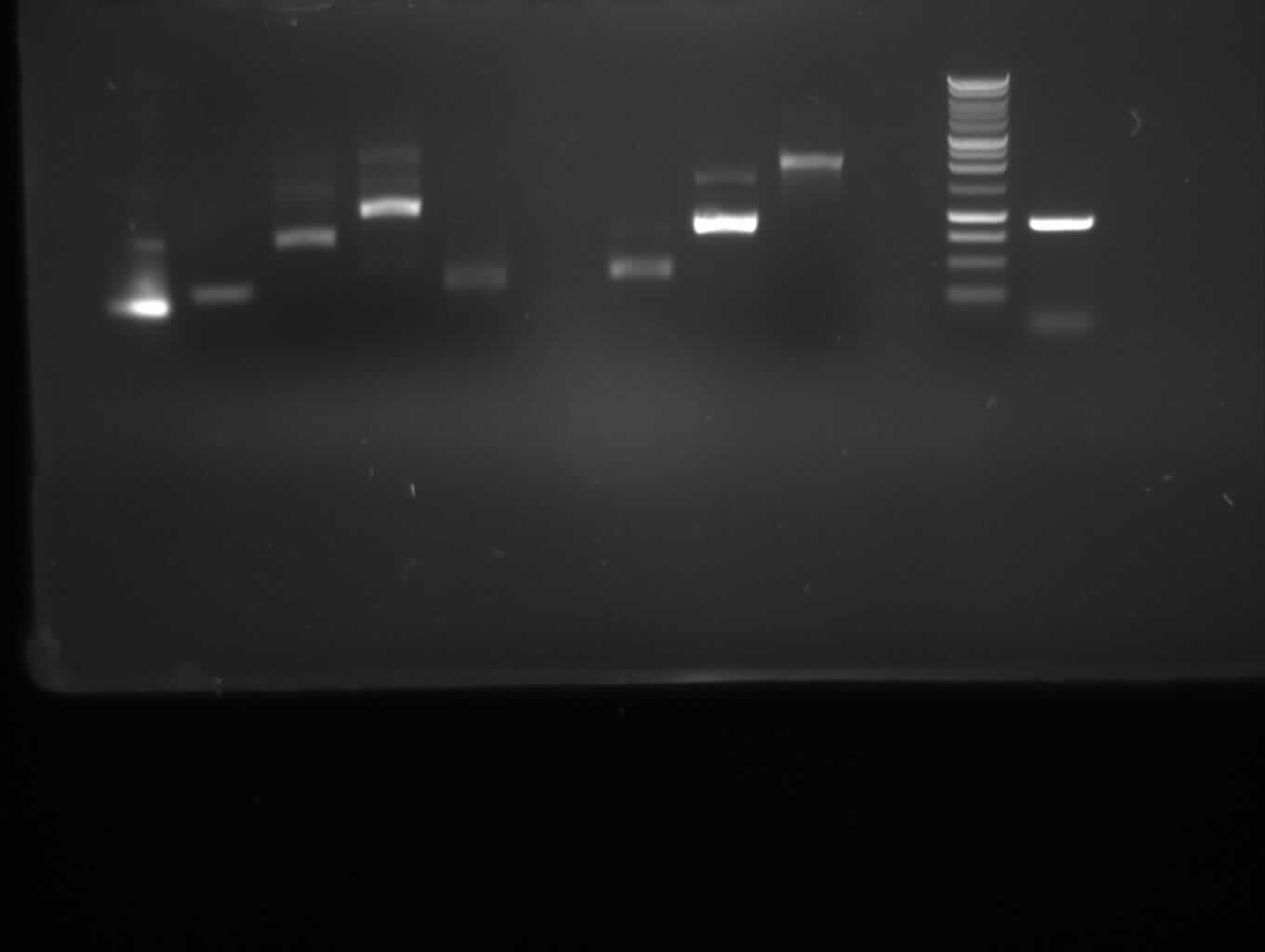
from left to right: PCR 1,2a,2b,3,4a,/,7b,9,10,/,ladder,pathway
| PCR nr.
| 1
| 2a
| 2b
| 3
| 4a
| 7b
| 9
| 10
|
| expected band (bp)
| 492
| 332
| 772
| 1087
| 330
| 402
| 808
| 1888
|
| shown band(s)
| 550,200
| 300
| 750
| 1100
| 300
| 450
| 900,1500
| 1900
|
| clean charge
|
| x
| x
| ~x
| x
| x
|
| x
|
new PCR for PCR6
PCR mixture for PCR6
| Concentration
| Low
| High
|
| template
| 1 µl (1:100)
| 15µl (1:100)
|
| primer (11,12)
| 2.5 µl*2
|
|
| dNTPs
| 1 µl
|
|
| Pfu
| 0.5 µl
|
|
| 10xbuffer
| 5 µl
|
|
| DMSO
| 1,25µl
|
|
| H2O
| 36.25 µl
| 22.25 µl
|
| sum
| 50µl
|
|
-> Protocol: Thermal cycler program: Touch down, 62°C-52°C, 30 cycles by 55°C
9-08-2010
Restriction digestion of eGFP, PCR6, ccdBamp
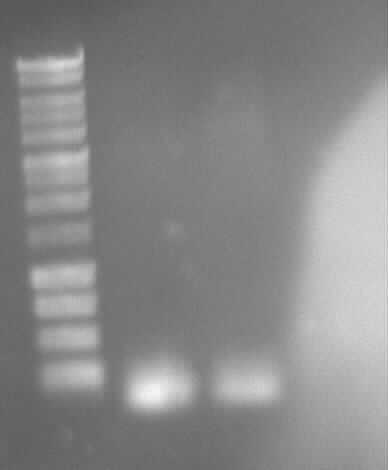 Gel photo of (left to right) PCR6(5ng/100ng) template as prepatation for ligation
-> Protocol 5 Restriction digestion
| eGFP
|
| SV40PA=PCR6
|
| ccdBamp
|
|
| DNA
| 5µl=300ng
| DNA
| 1µl=37,5ng
| DNA
| 2µl=115ng
|
| Buffer MC
| 2µl
| Buffer D
| 2µl
| Buffer H
| 2µl
|
| BSA 1:10
| 2µl
| BSA 1:10
| 2µl
| BSA 1:10
| 2µl
|
| EcoRI
| 0,5µl
| XbaI
| 0,5µl
| EcoRI
| 0,5µl
|
| SpeI
| 0,5µl
| PstI
| 0,5µl
| PstI
| 0,5µl
|
| H2O
| 13µl
| H2O
| 14µl
| H2O
| 13µl
|
| sum
| 23µl
| sum
| 20µl
| sum
| 20µl
|
1:30h 37°C, 20min 80°C
Agarose gel electrophoresis of PCR6
-> Protocol: 11 Agarose gel electrophoresis
25min, 150V
Ligation 1
| eGFP
| 14,4µL
| (144ng)
|
| SV40PA
| 4,8µL
| (48ng)
|
| ccdBamp
| 2,9µL
| (100ng)
|
| T4 Buffer 10x
| 3µL
|
|
| T4 Ligase
| 0,5µL
|
|
| H2O
| 4,4µL
|
|
| sum
| 30µL
|
|
Ligation at 22,5°C for 30 min, denaturation at 65°C for 10 min.
Concentration of DNA in Ligation 1:
Transformation
-> Protocol 18 competent cells2
The incubation time for the cells is here 1 hour.
9-09-2010
Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9
Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9
-> protocol 11 Agarose gel electrophoresis
results: "primer-clouds", PCR 9: no band
Plasmid extraction of ccdBcam, ccdBamp, Bak, CMV, PhiC31o
-> Protocol 4 Plasmid extraction from cells
| Plasmid
| concentration [ng/µL]
| A260/A280
|
| ccdBcam
| 22.5
| 1.0
|
| ccdBamp
| 42.5
| 1.2
|
| Bak
| 17.5
| 0.8
|
| CMV
| 10.0
| 0.7
|
| PhiC310
| 60
| 1.4
|
Eluated with H2O instead of the Eluation Buffer.
New PCR 1, 3, 4a, 5, 7a, 9, 10 with phusion
| PCRnr.
| 1
| 3
| 4a
| 4b
| 5
| 7a
| 9
| 10
|
| template
| pDS7 (1:100); 4µl
| 2a; 0.9µl+2b; 0.6µl
| Bak; 0.5µl
| Bak; 0.5µl
| 4a; 0.8µl+4b; 1µl
| 190-6 (1:100); 1µl
| eGFP (1:25); 2µl
| PhiC31o (1:10); 1µl
|
| primer [10pmol/µl]
| 1,2; 2.5µl
| 3,6; 2.5µl
| 7,8; 2.5µl
| 9,10; 2.5µl
| 7,10; 2.5µl
| 13,14; 2.5µl
| 20,21; 2.5µl
| 22,23; 2.5µl
|
| H2O
| 28µl
| 30.5µl
| 31.5µl
| 31.5µl
| 30.2µl
| 31µl
| 30µl
| 31µl
|
| Annealing temperature
| 51.5°C
| 53.1°C
| 53.1°C
| 51.5°C
| 53.1°C
| 57.5°C
| 61°C
| 57.5°C
|
+ in each assay:
dNTP mix: 1µl, 5x Phusion buffer: 10µl, DMSO:1.5µl, Phusion:0.5µl
sum: 50µl
Program: standard Phusion PCR, 29 cycles; anneling temperatures: see above
-> Protocol: 15 PCR with Phusion
9-10-2010
Agarose gel electrophoresis of PCR 1, 4a, 4b, 5, 7a
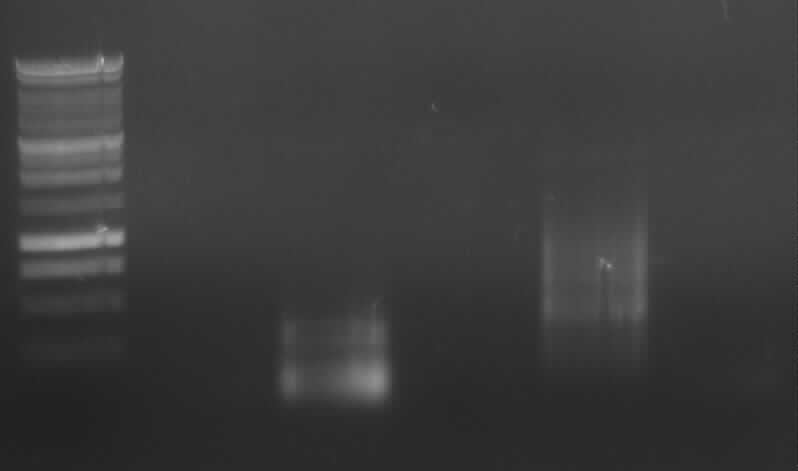
[From the left to the right: Ladder, 1, 4a, 4b, 5, 7a]
Only 4a has been amplified successfully.
Agarose gel extraction of PCR 3, 4a, 5, 9, 10
9-11-2010
weekend
9-12-2010
weekend
9-13-2010
text
9-14-2010
text
9-15-2010
text
9-16-2010
text
9-17-2010
text
9-18-2010
weekend
9-19-2010
weekend
9-20-2010
text
9-21-2010
text
9-22-2010
text
9-23-2010
text
9-24-2010
text
9-25-2010
weekend
9-26-2010
weekend
9-27-2010
text
9-28-2010
text
9-29-2010
text
9-30-2010
text
10-01-2010
text
10-02-2010
weekend
10-03-2010
weekend
|


![]()
![]()







![]()
![]()

![]()



![]()
![]()

 "
"















