Team:LMU-Munich/Jump-or-die/Schedule
From 2010.igem.org
(→Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequenz) |
(→Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequenz) |
||
| Line 60: | Line 60: | ||
==Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequenz == | ==Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequenz == | ||
| - | PCR1: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR1| | + | PCR1: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR1| Sequence of PCR1 from sequencing]]: confirmed [ ] |
| - | PCR5: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR5| | + | PCR5: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR5| Sequence of PCR5 from sequencing]]: confirmed [ ] |
| - | PCR6: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR6| | + | PCR6: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR6| Sequence of PCR6 from sequencing]]: confirmed [ ] |
| - | PCR9: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR9| | + | PCR9: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR9| Sequenceof PCR9 from sequencing]]: confirmed [ ] |
| - | PCR10: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR10| | + | PCR10: Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR10| Sequence of PCR10 from sequencing]]: confirmed [ ] |
==Aim 3: Assembling Biobricks== | ==Aim 3: Assembling Biobricks== | ||
Revision as of 10:37, 21 September 2010
For PCR key see PCR key
Colourcode for the primers: Annealing part, mutagenised part, other sequences integrated into primer
Primer used: 1TreF 2TreR
expected product size: 492 bp
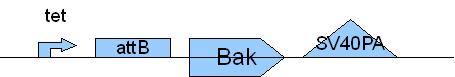
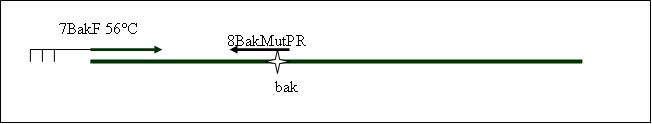
Primer used: 7BakF 8BakMutPR
expected product size: 330 bp
Primer used: 9BakMutPF 10BakR
expected product size: 376 bp
Primer used: 7BakF 10BakR
expected product size: 688 bp
Primer used: 11PAF 12PAR
expected product size: 237 bp
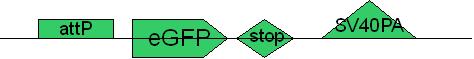
Primer used: 20eGFPattPF 21eGFPR
expected product size: 808 bp
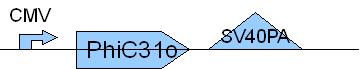
Primer used: 22PC31oF 23PC31oR
expected product size: 1888 bp
Note: for attB two Primers ( 18attBF 19attBR) used.
PCR1: Insertion [ ] -> Sequence of PCR1 from sequencing: confirmed [ ]
PCR5: Insertion [ ] -> Sequence of PCR5 from sequencing: confirmed [ ]
PCR6: Insertion [ ] -> Sequence of PCR6 from sequencing: confirmed [ ]
PCR9: Insertion [ ] -> Sequenceof PCR9 from sequencing: confirmed [ ]
PCR10: Insertion [ ] -> Sequence of PCR10 from sequencing: confirmed [ ]
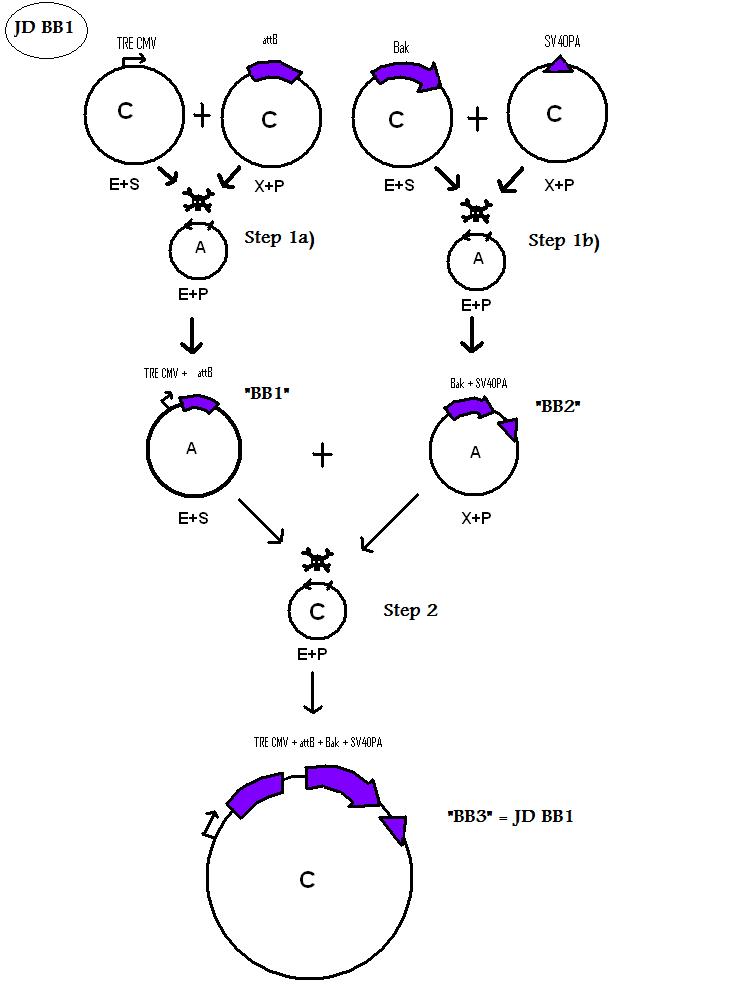
We are using the 3A System to assemble Biobricks.
Assembling Construct 1
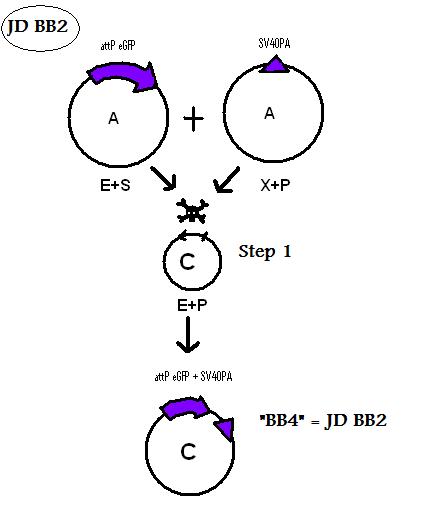
Assembling Construct 2
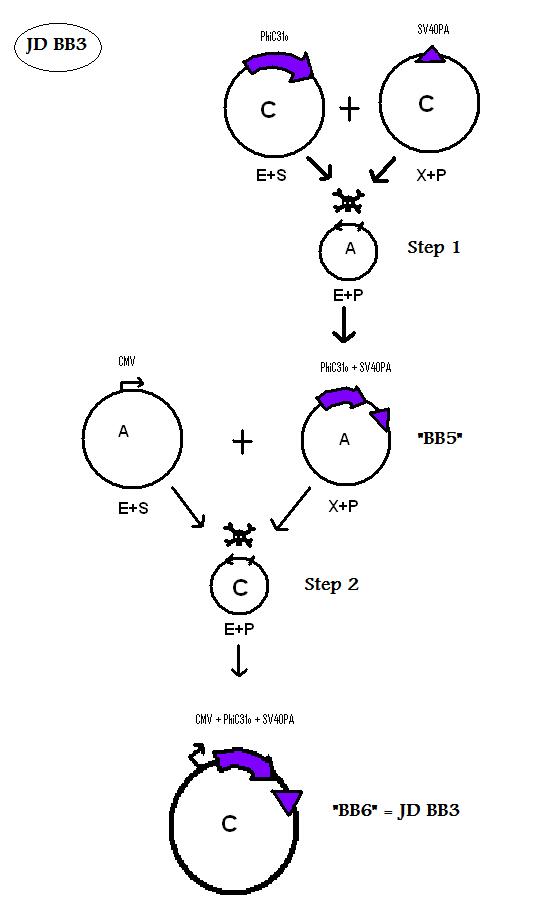
Assembling Construct 3
- Transform into HeLa cells to see if they survive.
-> Check the leakiness of tet-on-promoter
- Induce tet-on-promoter to see, if cells die.
-> Check if construct 1 is working
- Sequencing
- Transform into HeLa cells
- Western Blot or Proteinchromatography
-> Check if PhiC31o is read off


![]()
![]()







![]()
![]()

![]()
Contents
Aim 1: DNA reproduction+PCR
PCR1: replication of the Tet-inducible CMV promotor [X]

PCR4a: replication human bak with mutagenesis (Pst1) [ ]
PCR4b: replication human bak with mutagenesis (Pst1) [ ]
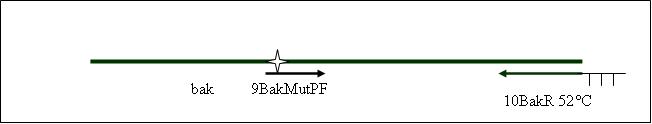
PCR5: joining PCR of human bak [ ]
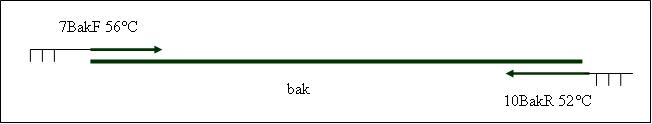
PCR6: replication of the SV40-polyadenylation site [X]

PCR9: replication of eGFP with attP in primer [ ]
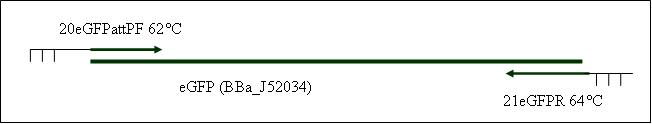
PCR10: Replikation of PhiC31o [X]

Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequenz
Aim 3: Assembling Biobricks
Aim 4: Testing products
Construct 1
Construct 2
Construct 3
Aim 5: Testing system
![]()
![]()

 "
"