Team:LMU-Munich/Cut'N'survive/Functional Principle
From 2010.igem.org
(Difference between revisions)
(→Construct 1) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| + | |||
| + | [[Image:cnsZelle.png|300px|Logo|right]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
== Cut'N'Survive-System == | == Cut'N'Survive-System == | ||
| Line 18: | Line 24: | ||
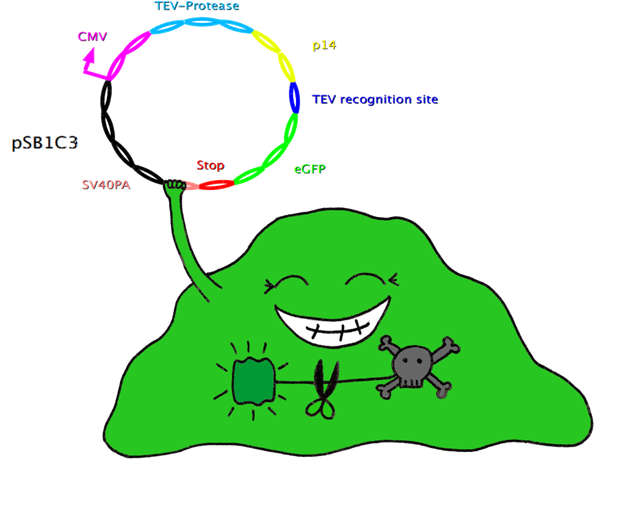
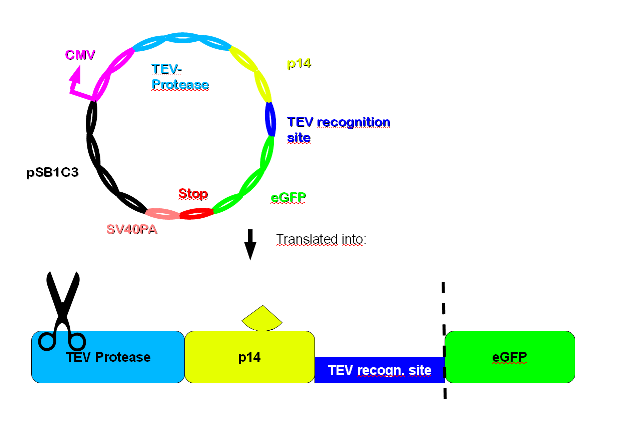
Construct 2 is composed of CMV-promoter, TEV-protease, p14, TEV-recognition site, eGFP, a double stop codon and SV40PA. | Construct 2 is composed of CMV-promoter, TEV-protease, p14, TEV-recognition site, eGFP, a double stop codon and SV40PA. | ||
| - | [[Image: | + | [[Image:cutwik.jpg|600pxs|TEVdegron-System: TEV-degron-System]] |
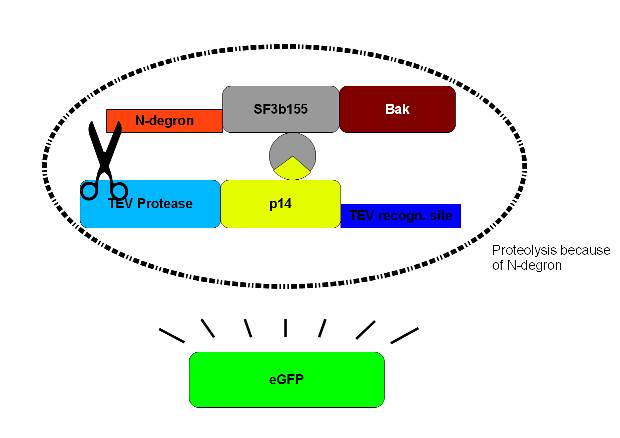
The CMV-promoter is for the expression of construct 2. The task of TEV-protease is to cut TEV-recognition sites, both the one upstream of N-degron to trigger protein degradation and the one upstream of the target gene (eGFP in this case) to ensure that the wanted product is not degraded. p14* is interacting with SF3b155 and so increases the rate of specific digestion at the TEV-recognition site by the TEV-proteases. eGFP is an example of a gene of interest which can conveniently be verified by green fluorescence. | The CMV-promoter is for the expression of construct 2. The task of TEV-protease is to cut TEV-recognition sites, both the one upstream of N-degron to trigger protein degradation and the one upstream of the target gene (eGFP in this case) to ensure that the wanted product is not degraded. p14* is interacting with SF3b155 and so increases the rate of specific digestion at the TEV-recognition site by the TEV-proteases. eGFP is an example of a gene of interest which can conveniently be verified by green fluorescence. | ||
| Line 38: | Line 44: | ||
==== Case b) cells incorporated with construct 2 ==== | ==== Case b) cells incorporated with construct 2 ==== | ||
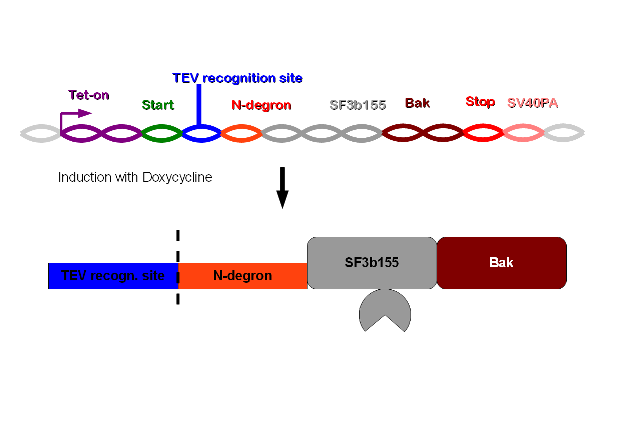
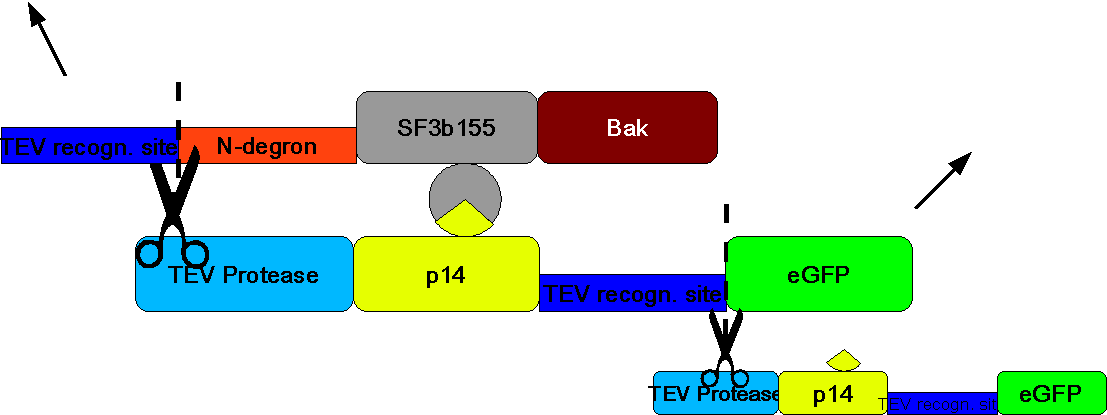
| - | Construct 2 will be read off and after introduction of | + | Construct 2 will be read off and after introduction of doxycycline the tet-on promoter will trigger the translation of construct 1 into protein. However in this case the TEV proteases will instantly separate eGFP from the rest of the protein as well as free the N-terminus of N-degron. This will cause the degradation of the whole protein complex - including bak. Therefore the cell will survive and only the protein product of the gene of interest will be left. |
| - | [[Image: | + | [[Image:cutwiki4.jpg|600px|TEVdegron-System: TEV-degron-System]] |
| - | [[Image:cutwik5.jpg| | + | [[Image:cutwik5.jpg|600px|TEVdegron-System: TEV-degron-System]] |
| + | [[Image:cnsZelle.png|300px|Logo|right]] | ||
Latest revision as of 14:39, 25 October 2010


![]()
![]()







![]()
 "
"