Team:KAIST-Korea/Project/StructureAlignment
From 2010.igem.org
|
Structure Alignment
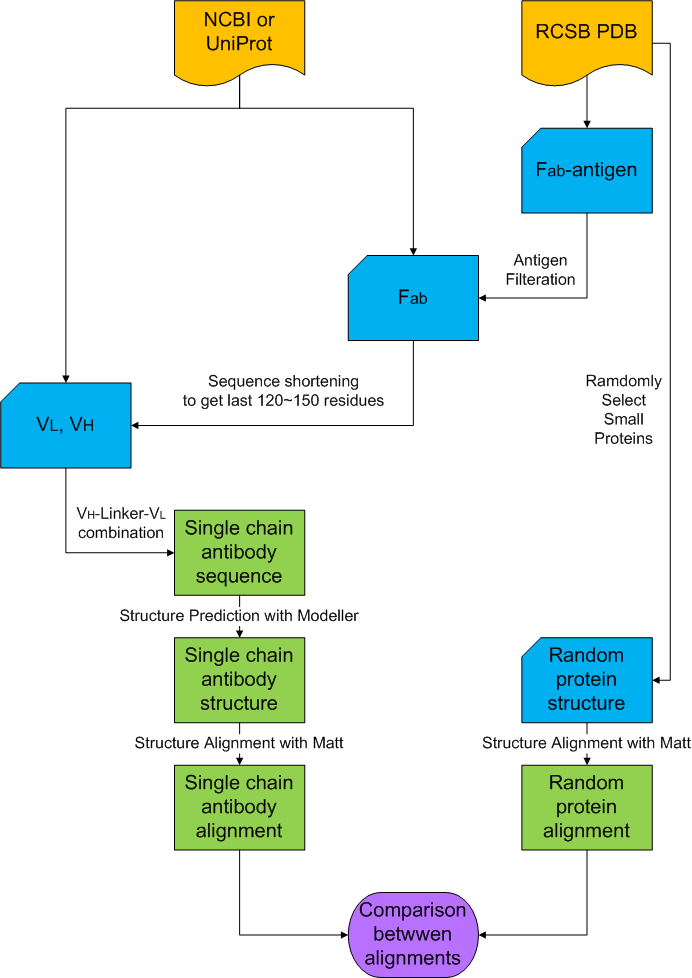
Single chain antibody structural alignmentProtocolThere are four steps to compare structure of single chain antibody and FGF binding domain of FGFR. The first step is to take variable region sequences of antibodies. The next one is to combine these variable region sequences with a linker sequence to make single chain antibody sequence. Then, we predict the structure of single chain antibody with a structure prediction program like Modeller. Lastly, we structurally align these structures of antibodies with structure of FGF binding domain of FGFR (PDB ID: 1EVT).
Data sourceSingle chain antibody is the combination of variable regions of known antibodies with linker sequence, which can bind to the antigens. We need to know the VL and VH sequences to make single chain antibody. The sources of these antibody sequences include NCBI, Uniprot and RCSB PDB. NCBI and Uniprot provide the single chain sequence of variable regions (VL and VH) and antigen binding fragments (Fab). RCSB provide the structure of antigen binding fragment complexes that bind to their antigens. We, however, only need the sequence of variable region. So, we get the last 120~150 residues and assume them as the variable region. And, data from RCSB contain not only sequence of antibody but also antigens. Therefore, we filter them based on label of files to get heavy chains and light chains of antibody.
Single chain antibody synthesisWe combine the antibody variable region sequences in order of VH-linker-VL to make single chain antibody sequence. The sequence of the linker is GGGGSGGGGS.
Structure PredictionWe used the program called Modeller to predict the structure of single chain antibody from its sequence. Modeller predicts 3D structure of protein with structure of known similar proteins based on homology model. Input file is the sequence of single chain antibody in fasta format and output file is the structure of single chain antibody in pdb format.
Structure AlignmentIn this step, we check the structural similarity between single chain antibodies and FGF binding domain to align the structure of single chain antibody with that of FGF binding domain of FGFR. The structure of FGF binding domain of FGFR is provided by RCSB PDB (PDB ID: 1EVT). We used Matt structural alignment program to do this job. Matt performs the structural alignment, which minimizes the distance between α-carbon chains of two proteins based on the common structure (α helix). Input file is the structure of single chain antibody in pdb format and output file is a text file that contains the number of amino acids, which are composed of shared structure (Core residue), average distance between alpha carbon chains of two proteins (Core RMSD), the score of similarity, which is calculated by Matt (Raw score), and the probability that this similarity is just a product of random (p-value) and pdb files, which contain the alignment result of single chain antibody with FGF binding domain of FGFR.
Result : Table
Result : FiguresUsing arranged pdb files and protein 3D structure drawing program - PyMOL, again, we predict structural similarity between the structure of FGF binding domain and that of single chain antibodies associated with the table. In order to show the structures clearly, we control the shape setting 'cartoon' and 'chain'. Cyan Color represents the structure of FGF binding domain and Green Color is for the comparative antibodies. In the result, we can easily see all antibodies' structures are very similar, completely or symmetrically.
  Result AnalysisThe table above shows the result of alignments of 65 single chain antibodies for control group and 1 single chain antibody for our real experiment (16A1). Higher core residue and Raw score, lower core RMSD, and p-value mean more similarity. 16A1 antibody shows 26th highest in core residue, 2nd lowest in core RMSD, 13th highest Raw score and 13th lowest p-value. These result means that our 16A1 antibody has higher similarity than average antibodies have. In general, Core RMSD value of randomly selected small protein (colored brown) is lower than that of single chain antibody (colored green). It may seem confusing, but lower Core RMSD is caused by RMSD calculation of Matt. Matt calculates RMSD for not whole proteins, but only shared structures. Therefore, we should check if protein pairs with lower Core RMSD have enough number of Core residues. |
 "
"