Team:KAIST-Korea/FutureWorks
From 2010.igem.org
Luftschloss (Talk | contribs) (→Life Cycle of S.pombe) |
Luftschloss (Talk | contribs) (→Life Cycle of S.pombe) |
||
| Line 25: | Line 25: | ||
=== Life Cycle of ''S.pombe'' === | === Life Cycle of ''S.pombe'' === | ||
[[Image:LifeCycleOfSpombe.jpg|right]] | [[Image:LifeCycleOfSpombe.jpg|right]] | ||
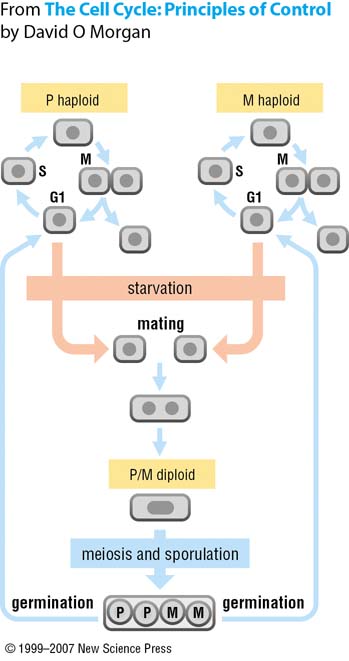
| - | Before description of our idea, we should talk about the life cycle of ''S.pombe'' | + | Before description of our idea, we should talk about the life cycle of ''S.pombe''. Generally, ''S.pombe'' is haploid organism. So they have only one copy of each gene. But in starving condition, Cells of ''S.pombe'' become diploid through mating. And this mating allow yeast have the gene from different parents. Therefore, with the starvation, we can induce the conjugation of ''S.pombe'' and integrate genes from two different gene sets. |
=== How to implement? === | === How to implement? === | ||
Revision as of 11:29, 27 October 2010
|
Future Works
Our Final Goal and Problem of Cross-talking implementationOur final goal is to develop the universal diagnosis kit for every disease whose antibody is produced. To perform this goal, the yeast should express modified antigen receptor of wanted disease, STAT1 proteins and have GFP gene whose promoter have GAS element. Most easiest way to way to detect existence of antigen is to express every antigen receptor on one yeast cell and integrate these signal into GFP with common STAT1 dimerization pathway. But this way have problems. Though the GFP is expressed, we can't sure which antigen is detected, because any antigen binding can turn on the GFP expression. Therefore, the cross-talking implementation can't be used for differential diagnosis because it can't eliminate any disease which can turn on the GFP expression. Disadvantage of All-in-One ImplementationSo we tried to selectively express modified antigen receptor genes to use promoter with different activator molecule. With this All-in-One implementation, one yeast cell have genes for GFP with GAS element, STAT1 signal transduction molecule and every modified antigen receptor with promoters whose activator is different. In theoretically, this implementation solve the problem of cross-talking of modified antigen receptors. But this implementation have two problems.
Our Final Implementation : ConjugationSo we gave up to insert genes for every modified antigen receptors. But we still wanted to make universal diagnosis kit. So we looked for the way to 'combine the genes'. And we found the 'conjugation of S.pombe' With certain condition, If spores from different yeast can be mated, their genes can be in same cell. Therefore, we divide our diagnosis system to specific gene and common genes. And each set of genes are inserted into different yeast. And the diagnostician would make the conjugated cell with whole sensing pathway to stimulate the mating of yeast spore. Life Cycle of S.pombeBefore description of our idea, we should talk about the life cycle of S.pombe. Generally, S.pombe is haploid organism. So they have only one copy of each gene. But in starving condition, Cells of S.pombe become diploid through mating. And this mating allow yeast have the gene from different parents. Therefore, with the starvation, we can induce the conjugation of S.pombe and integrate genes from two different gene sets. How to implement? |
 "
"