Team:Johns Hopkins/Project
From 2010.igem.org
Arjunkhakhar (Talk | contribs) (→Instrumentation) |
Arjunkhakhar (Talk | contribs) (→Visualizing the Crz1 Transcription Factor) |
||
| Line 62: | Line 62: | ||
<object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> | <object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> | ||
</html><br> | </html><br> | ||
| - | In this video we can see the GFP tagged | + | In this video we can see the GFP tagged CRZ1 cells displaying nuclear localization after shocking. The results can be seen only in 4-5 cells in the video due to an inefficient electro stimulation platform being used at the time. We did, however, see repeatable results of nuclear localization. |
===Optimizing Parameters for the CDRE from the FKS2 Promoter=== | ===Optimizing Parameters for the CDRE from the FKS2 Promoter=== | ||
Revision as of 23:54, 25 October 2010
Contents |
Abstract
If the goal of iGEM and the Parts Registry is to take the messy world of genetic engineering and transform it into something like the standardized world of electrical engineering, it may be useful if electronic systems could directly interface with biological systems. Past iGEM projects have used chemical or optical stimuli to actuate transcriptional responses. Our project, however, seeks to add voltage sensitivity to Saccharomyces cerevisiae. Baker’s yeast was chosen because in some sense yeast have a system that responds to voltage input. With a voltage stimulus one can open the voltage-gated calcium channels of yeast, causing calcium ions to rush into the cytoplasm. This causes calcineurin to dephosphorylate Crz1, which enters the nucleus and binds various promoters. Our group presents a library of characterized Crz1-sensitive promoters of both naturally-occurring and synthetic varieties. Genes downstream of these promoters are thus voltage-regulated in media containing calcium.
Aims
- Show that voltage can be used to stimulate a transcriptional response in S. cerevisiae.
- Develop a library of voltage-inducible promoters with differing voltage response curves.
- Determine the functional range of and optimized values for our system with respect to the following variables:
- Voltage applied
- Duration of voltage application
- Presence of vacuoles, yeast’s natural mode of intracellular Calcium control
- Develop an effective experimental apparatus to apply voltage and measure response.
Methods
Visualizing the Crz1 Transcription Factor
We want to characterize the electro stimulation voltage domain in which we see movement of the GFP tagged CRZ1 transcription factor into the nucleus. Crz1-GFP was grown and passed into two rows of a 96 well plate. The cells were then shocked every two hours at 10 Volts. The cells were passed to a glass plate where they were fixed and then imaged. GFP was observed to move in and out of the nucleus just 5 minutes after voltage stimulus. In a given image, some cells displayed GFP densely packed in the nucleus while other cells displayed GFP in the cytoplasm, but excluded from the nucleus. Here we see a confirmation of the oscillating behavior described by Elowitz et al (Elowitz 2008).
In this video we can see the GFP tagged CRZ1 cells displaying nuclear localization after shocking. The results can be seen only in 4-5 cells in the video due to an inefficient electro stimulation platform being used at the time. We did, however, see repeatable results of nuclear localization.
Optimizing Parameters for the CDRE from the FKS2 Promoter
We want to find an optimum voltage amplitude and electro stimulation time for yeast containing the CDRE-RFP plasmid. Shocking at 10 Volts caused large amount of cell death, as such, it was necessary to find an optimal voltage to cause transcription, but not to damage our cells. CDRE-mCherry were grown and passed into 96 well plate. The electroporator was used to shock the cell with voltages from 2-10 Volts with an exposure time from 0-80 seconds. We find optimum electro stimulation voltage is 8V and we need at least 40 seconds of electrostimulation to see expression.
| 2s | 5s | 10s | 20s | 40s | 80s | |
|---|---|---|---|---|---|---|
| 10V | None | None | None | None | None | N/A |
| 8V | None | None | None | None | High | High |
| 6V | None | None | None | None | Moderate | High |
| 4V | None | None | None | None | Low | Low |
Further Honing FKS2 CDRE
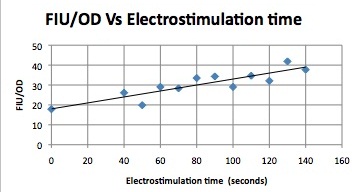
We want to hone in on the FSK2-CDRE systems region of activation in the domain of time of shocking. Also to compare yeast without PMC1 and VCX1 knocked out versus yeast with it not knocked out to determine effect of the knockout on the CDRE systems expression levels. From here we see a through characterization of the system that shows increase in expression of our reporter RFP with increase in electro stimulation time. We also see that there is much higher expression of the cells without vacuoles knocked out than in cells with vacuoles knocked out.
FKS2 CDRE and PMC1 CDRE in Vacuole Positive and Negative Yeast
A large amount of cells were dying from the stresses of voltage shocking, as such, a FKS2 CDRE-mCherry was inserted into yeast that still had their calcium vacuoles. Yeast were grown and passed into two rows of a 96 well plate and shocked at 2-10 volts with an exposure time of 0-80 seconds. We want to characterize the relationship between electro stimulation times and expression of florescent protein of the synthetic CDRE promoter sequence and the PMC1 promoter sequence. We also want to demonstrate the effect of knocking out the PMC1 and VCX1 genes in yeast.
Instrumentation
We tried several approaches to efficiently and accurate electro stimulate our cells with Killing them:
- A Micro fluidic chip with 300um wide channels with electrodes to shock cell with. We did manage to build it, however, due time constraints we were not able to adequately test it to optimize its use.
- A 8 Well plate with aluminum electrodes which had a brass backing. This did show us our first results, but due to the inaccuracy of the instrument we had to abandon its use.
- Finally we have started using a gold plated coaxial 8 well electroporater to electrically stimulate the cells. It presents some problems as far as modeling the field effect on the cells goes, but it has shown excellent results and it allows us to do large scale experimentation using 96 well plates.
PICTURES
Future Research Plans
Our future research plans are to finish categorizing the synthetic CDRE sequences in our promoter sequence using our high throughput electro stimulation device. We also plan to run a complete set of trials of the CDRE system on our microfluidic electro stimulation platform, once the optimization for the same is complete, to be able to a more through set of results in a more controllable environment. We also plan to apply our optimized high throughput experimentation technique to a library of synthetic CDRE sequences we have hypothesized to characterize their voltage activation domains. LETS MENTION LIBRARY NOAH GENERATED WITH PROBABILITES HERE Finally we hope to integrate a repressor based system involving 2 CDRE controlled genes to demonstrate the several levels of control that be achieved with this system.
References
- Long Cai, Chiraj K. Dalal & Michael B. Elowitz. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature (2008).
 "
"