Team:Imperial College London/Results/Exp7
From 2010.igem.org
(New page: {{:Team:Imperial_College_London/Templates/Header}} {{:Team:Imperial_College_London/Templates/ResultsHeader}} {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helve...) |
|||
| Line 11: | Line 11: | ||
The current method of characterizing expression systems is the one introduced by Jason Kelly et al. (2009), Measuring the activity of BioBrick promoters using an in vivo reference standard. Citing from the actual publication conclusion section: “Relative activity measurements based on an in vivo reference standard enables improved measurement of promoter activity given variation in measurement conditions and instruments. These improvements are sufficient to begin to support the measurement of promoter activities across many laboratories”. Using this method we decided to test one of our key parts by placing the XylE gene under the influence of two different promoters; the J23101 promoter and the pVeg promoter. | The current method of characterizing expression systems is the one introduced by Jason Kelly et al. (2009), Measuring the activity of BioBrick promoters using an in vivo reference standard. Citing from the actual publication conclusion section: “Relative activity measurements based on an in vivo reference standard enables improved measurement of promoter activity given variation in measurement conditions and instruments. These improvements are sufficient to begin to support the measurement of promoter activities across many laboratories”. Using this method we decided to test one of our key parts by placing the XylE gene under the influence of two different promoters; the J23101 promoter and the pVeg promoter. | ||
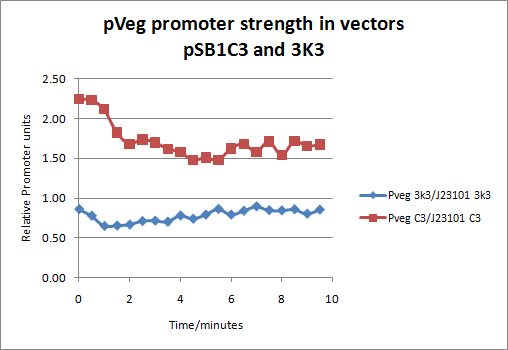
| - | Our experiment was performed on 96-well plates comparing four different populations of Catechol(2,3)dioxygenase expessing E.coli cells. These four different populations were: 1) XylE under the influence of J23101 promoter in pSB1C3 vector, 2) ) XylE under the influence of J23101 promoter in 3K3 vector, 3) XylE under the influence of pVeg promoter in pSB1C3 vector and 4) ) XylE under the influence of pVeg promoter in 3K3 vector. All 4 populations were grown under same condition and tested for the expression of catechol dioxygenase enzyme by addition 0.25mM catechol substrate. Reaction was monitored using spectrophotomer and the | + | Our experiment was performed on 96-well plates comparing four different populations of Catechol(2,3)dioxygenase expessing E.coli cells. These four different populations (initial O.D. 0.5) were: 1) XylE under the influence of J23101 promoter in pSB1C3 vector, 2) ) XylE under the influence of J23101 promoter in 3K3 vector, 3) XylE under the influence of pVeg promoter in pSB1C3 vector and 4) ) XylE under the influence of pVeg promoter in 3K3 vector. All 4 populations were grown under same condition and tested for the expression of catechol dioxygenase enzyme by addition 0.25mM catechol substrate. Reaction was monitored using spectrophotomer and the production of colored product was recorded. Production of HMS colored product is directly proportional to total enzyme presence. Since total enzyme presence depends on the strength of the promoter, this provides a means for quantification of the strength of promoters in relative promoter units. |
| + | |||
| + | The results showed that pVeg promoter in pSB1C3 vector, a high copy plasmid, has an 1.62 r.p.u value and in 3K3 vector, a low copy plasmid an 0.79 r.p.u. value. These values were derived by dividing signal from the production of HMS by the pVeg promoter population of cells by signal from the standard promoter J23101 (r.p.u value of 1).Figure below: | ||
| + | [[Image:Untitled 5.jpg|thumb|center|400px|Graph showing the ratio of pVeg/J23101 signals in pSB1C3 and 3K3 vectors]] | ||
|} | |} | ||
Revision as of 23:36, 27 October 2010
| Experimental Results | Exp 1 | Exp 2 | Exp 3 | Exp 4 | Exp 5 | Exp 6 | Exp 7 |
| Testing is a fundamental stage of the engineering design cycle and is a crucial part of charactrising BioBrick Standard Biological Parts so that other people can benefit from our work. We've compiled all our results on this page, detailing how the experiments were carried out and the significance of the data. | |
| Experiment 7 | Characterization in r.p.u. of Pveg promoter | |
|
The notion of using Catechol dioxygenase as a reporter gene for characterizing expression circuits was put forward in previous experiments performed by our lab team. Current reporter genes, although some of them well established in the biosciences community, are not ideal for systems that require special conditions. The frequently used and widely accepted GFP protein would not be ideal for use in experimentation with thermophilic bacteria, where it does not fold properly, or systems where GFP is incorporated into low copy loci such as chromosome, an F factor or low copy plasmid. As an alternative, catechol dioxygenase would be ideal for these systems as it folds properly in higher temperatures and gives a detectable signal even in very low numbers of expression. Another advantage of this reporter enzyme is that its output signal is a colored product detectable by a spectrophotometer, a pretty standard equipement in all labs nowadays. The current method of characterizing expression systems is the one introduced by Jason Kelly et al. (2009), Measuring the activity of BioBrick promoters using an in vivo reference standard. Citing from the actual publication conclusion section: “Relative activity measurements based on an in vivo reference standard enables improved measurement of promoter activity given variation in measurement conditions and instruments. These improvements are sufficient to begin to support the measurement of promoter activities across many laboratories”. Using this method we decided to test one of our key parts by placing the XylE gene under the influence of two different promoters; the J23101 promoter and the pVeg promoter. Our experiment was performed on 96-well plates comparing four different populations of Catechol(2,3)dioxygenase expessing E.coli cells. These four different populations (initial O.D. 0.5) were: 1) XylE under the influence of J23101 promoter in pSB1C3 vector, 2) ) XylE under the influence of J23101 promoter in 3K3 vector, 3) XylE under the influence of pVeg promoter in pSB1C3 vector and 4) ) XylE under the influence of pVeg promoter in 3K3 vector. All 4 populations were grown under same condition and tested for the expression of catechol dioxygenase enzyme by addition 0.25mM catechol substrate. Reaction was monitored using spectrophotomer and the production of colored product was recorded. Production of HMS colored product is directly proportional to total enzyme presence. Since total enzyme presence depends on the strength of the promoter, this provides a means for quantification of the strength of promoters in relative promoter units. The results showed that pVeg promoter in pSB1C3 vector, a high copy plasmid, has an 1.62 r.p.u value and in 3K3 vector, a low copy plasmid an 0.79 r.p.u. value. These values were derived by dividing signal from the production of HMS by the pVeg promoter population of cells by signal from the standard promoter J23101 (r.p.u value of 1).Figure below: |
 "
"