Team:Imperial College London/Lab Diaries/XylE team
From 2010.igem.org
| Line 81: | Line 81: | ||
<li><a href="#Week_8" class="popt">Week 8</a></li> | <li><a href="#Week_8" class="popt">Week 8</a></li> | ||
<li><a href="#Week_9" class="popt">Week 9</a></li> | <li><a href="#Week_9" class="popt">Week 9</a></li> | ||
| + | <li><a href="#Week_10" class="popt">Week 10</a></li> | ||
| + | <li><a href="#Week_11" class="popt">Week 11</a></li> | ||
<li><a href="#Lab_Diary" class="popt">Lab Diary</a></li> | <li><a href="#Lab_Diary" class="popt">Lab Diary</a></li> | ||
<li><a href="#Output_Photo_Gallery" class="popt">Output Photo Gallery</a></li> | <li><a href="#Output_Photo_Gallery" class="popt">Output Photo Gallery</a></li> | ||
| Line 299: | Line 301: | ||
The transformation of E.Coli with PSB1-C3 with insert did not work :( | The transformation of E.Coli with PSB1-C3 with insert did not work :( | ||
|} | |} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | =='''Week 11''' == | ||
| + | |||
| + | [[Image:wk11.jpg|thumb|800px|13.9.10-17.9.10]] | ||
| + | |||
| + | ===Monday 13th september=== | ||
| + | Overnights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. | ||
| + | |||
| + | ===Tues 14th September=== | ||
| + | [[Image:CS1.JPG|thumb|right|300px|test confirming that yellow product of XylE enzmymatic reaction is leaks back from the cytosol into the solution.1C and 2C is the XylE producing cells after centrifuging and redissolving of the pellet and 1S and 2S is the supernatant after centrifuging]] | ||
| + | *assay on plate reader of: | ||
| + | **0-0.75 initial O.D. of transformed E.coli | ||
| + | **0-1mM initial catechol concentration | ||
| + | The assay was carried out with E.coli, top ten spcies, transformed with J23101-XylE-B0014 in pSB1C3 vector.The overnight culture wastransfered in new medium this morning for 4 hrs before assaying. LB medium was used for dilutions and blank. Catechol was diluted in ddH2O. | ||
| + | [[Image:Catechol assay test 2.xls|Data analysis of the assay]] | ||
| + | *Mini preps of XylE 8,10 colonies and 3K3;102 151 260 colonies. Analytical digests were performed using E+S on XylE, E+S AccI and HindIII+E for 3K3 vector respectively. The band patterns correctly identified the vector to be 3k3. Colony 10 of XylE and Colony 260 of the 3k3 vector were set up direct into 100ml LB and so will require til 12pm for midi prepping on weds. Nick performed a second plate reader assay to determine the minimal concentration of Catechol to use for assays. I took 990ul M9 culture containing colony 10 and 10ul catechol was added. This was incubated for 10 mins and then spun down. The supernatant was removed into a cuvette and the cells resuspended in M9 salts. The ODs were then read on the spectrophotometer at 380 and 600nm. It was found that control cuvette of M9 salts had a miniscule reading. M9 + cells ~0.007. Supernatant + cells were ~ 2.5 therefore we could deduce that the coloured catechol breakdown product is exported out of the cells. | ||
| + | |||
| + | Piotr: | ||
| + | *Mini prep of 1 sample (6 samples lost due to mistake) of GFP-Xyle fusion protein and its digestion with Spe and Xba (gel to be run on the next day) | ||
| + | *Preparing E.Coli colonies for the next day for mini prep to be redone | ||
| + | *PCR of 6 samples of GFP-Xyle fusion | ||
| + | |||
| + | ===Weds 15th September=== | ||
| + | |||
| + | *Midi preps of overnights. | ||
| + | *In preparation for assays determining the effect of catechol and/or breakdown product on cell viability we prepared Top10 cells, one strain containing pVeg-XylE-terminator and the other containing a CMR plasmid of similar size. As the XylE cells have already been prepared, only Top10 transformation with CMR-plasmid had to be carried out. | ||
| + | |||
| + | ===Thurs 16th September=== | ||
| + | *Top10 CMR transformation was successful. Four overnight cultures of each Top10 XylE and Top10 CMR were set off with either M9 or LB medium respectively. | ||
| + | *Piotr: Did mini preps from bacteria with blunt ended Xyle-GFP fusion and did diagnostic using both: | ||
| + | |||
| + | - digest with XbaI and SpeI | ||
| + | |||
| + | - PCR reactions with the following primers added: HIS-GFP, XylE-Xba and the other sample with HIS-GFP and GFP-flag | ||
| + | |||
| + | The PCR reactions were not very conclusive on the gels but digests allowed to determine that colonies 1->5 seem to have the right sizes of DNA in them and on Friday they will be prepared to be sent off for sequencing | ||
| + | |||
| + | *continuation of the midi from overnight step. A gel analysis was run which showed the correct bands were present. A gel purification of the protein was then performed and the bands excised. | ||
| + | |||
| + | ===Friday 17th September=== | ||
| + | *Catechol assay of transformed E.coli Top ten(J23101-XylE-terminator) in M9 medium + data analysis | ||
| + | **[[Image:M9 catechol concentrations assay alternative.xls|Data analysis]] | ||
| + | **[[Image:Catechol assay in M9.xls| alternative]] | ||
| + | |||
| + | The gel purifications of 3k3 vector and XylE were used for ligation. | ||
| + | 3K3 was dephosphorylated and then ligated in a ratio 5:1 with the insert. This was left overnight and Chris tried to transform with it on sunday.. ligation and transformation failed :-( | ||
| + | |] | ||
Revision as of 09:47, 21 October 2010
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
| Team XylE |
Follow our progress: Click me!
| XylE team Lab Objectives |
|
| Lab notes and schedule |
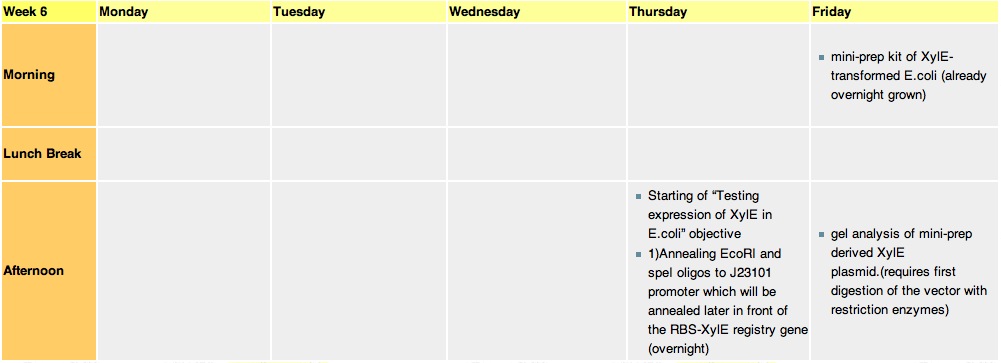
Week 6Thursday, 12-Aug-2010
we constructed the standard E.coli promoter J23101 with sticky ends. These ends are complementary to restriction sites made by EcoRI and SpeI enzyme. This promoter will be later used in 3A assemply to construct a promoter-RBS-XylE design in a psB1C3 vector. E.coli will be transformed with this final construct plasmid to assess XylE activity and characterization. It will also be one of the submitted biobricks.
these cultures are going to be used tomorrow for mini-prepping. Miniprep will allow us to isolate E.coli's plasmid DNA(which contains the XylE gene). Friday, 13-Aug-2010
Mini-prep is usually used to confirm that our gene of interest has not been changed in any way, as the isolated plasnid id sent for sequencing. However, since XylE was taken from the registry, we assume that it is fine and no sequencing is required. The mini-prep will later be used for the midi-prep (that gives out higher yeilds of DNA needed for cloning).
|
|
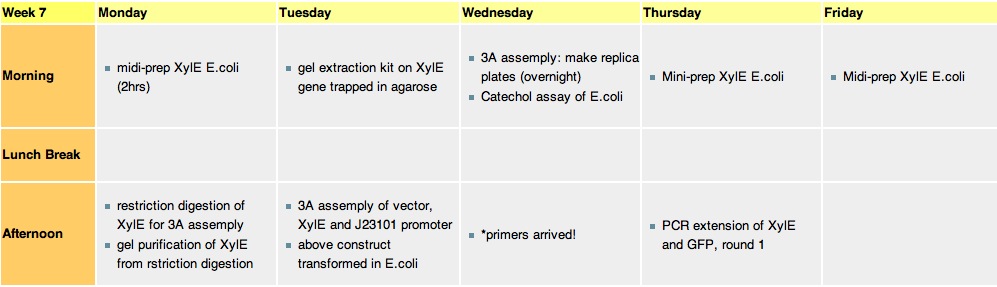
Week 7Monday, 16-Aug-2010
Tuesday, 17-Aug-2010
Thursday, 19-Aug-2010
Friday, 20-Aug-2010The J23101 gene in a biobrick vector containg RFP gene
|
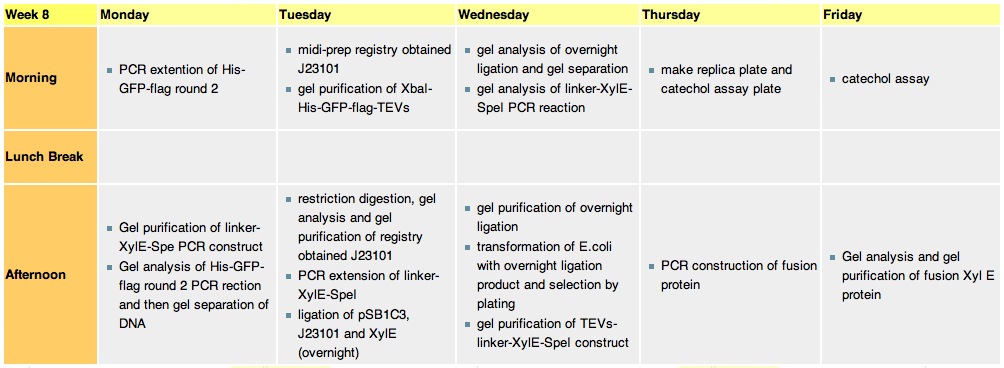
Week 8Monday, 23-Aug
Tuesday, 24th-Aug
Wednesday, 25th-AugPerformed gel analysis on the purified XylE and J23101 to obtain ratios for ligation. First gel was scrapped as it produced appauling(explanation for Nick:really bad) results, 2nd gel run was successful.
Thursday, 26th-Aug
Friday, 27th-Aug
File:Catechol Assay before.jpg Plate before adding catechol assay File:Catechol assay after (27-8).jpg After addition of catechol colonies turn yellow-orange in seconds!!
Saturday, 28th-Aug
Sunday, 29th-Aug
|
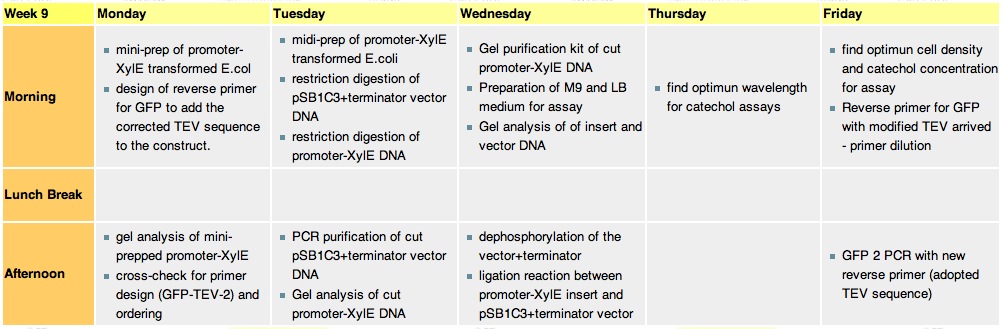
Week 9
Monday, 30th-Aug
Tuesday, 31st-Aug
Thursday, 2nd-SeptFile:200-600nm spectr.jpg Spectra of XylE transformed E.coli after addition of catechol assay. The broad peak around 380nm wavelength arises is due to the presence of the product of the enzymatic reaction involving pyrocatechol and XylE enzyme. This peak if absent if a culture of XylE transformed cells are measured without the addition of catechol
Friday, 3rd-Sept
File:Assay 3 sept.jpg Catechol assay on XylE-trasformed cells in a 96-well plate (A to H decreasing cell concentration, 1-10 decreasing catechol concentration, column 11 and 12 negative and control) |
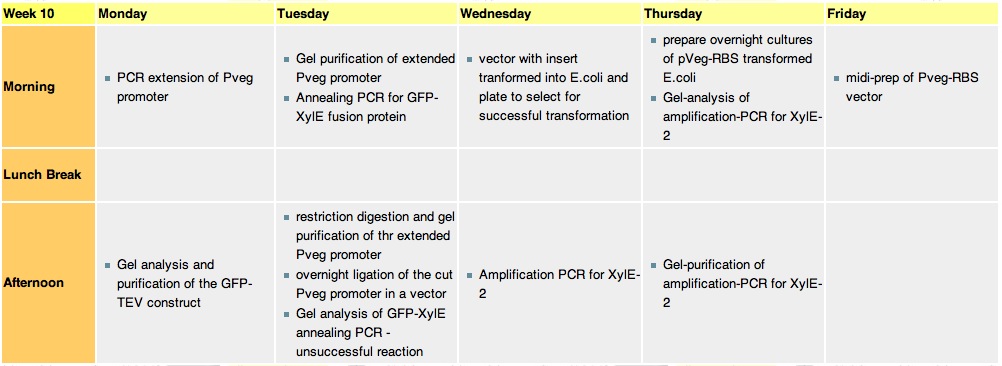
Week 10Monday, 6th-Sept
Tuesday 7th Sept
Wednesday, 8th Sep
Thursday, 9th Sep
Friday, 10 sepThe transformation was a SUCCESS. 2x replica plates were made plate 1# 1-6 plate #2 6-11; colony 6 and colony 9 of plates 1 and 2 respectively were transfered into a 5ml liquid culture + 5ul CmR. These will later be turned into glycerol stocks. After the replica plates have grown up mini preps on a number of colonies shall be performed - this hopefully will eliminate the contaminating plasmid DNA. This will be followed by a midi prep. Saturday, 11 SepThe transformation of E.Coli with PSB1-C3 with insert did not work :( |
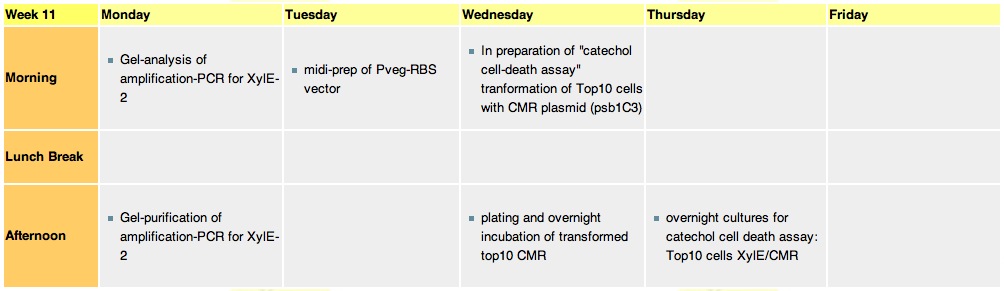
Week 11Monday 13th septemberOvernights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. Tues 14th September
The assay was carried out with E.coli, top ten spcies, transformed with J23101-XylE-B0014 in pSB1C3 vector.The overnight culture wastransfered in new medium this morning for 4 hrs before assaying. LB medium was used for dilutions and blank. Catechol was diluted in ddH2O. Data analysis of the assay
Piotr:
Weds 15th September
Thurs 16th September
- digest with XbaI and SpeI - PCR reactions with the following primers added: HIS-GFP, XylE-Xba and the other sample with HIS-GFP and GFP-flag The PCR reactions were not very conclusive on the gels but digests allowed to determine that colonies 1->5 seem to have the right sizes of DNA in them and on Friday they will be prepared to be sent off for sequencing
Friday 17th September
The gel purifications of 3k3 vector and XylE were used for ligation. 3K3 was dephosphorylated and then ligated in a ratio 5:1 with the insert. This was left overnight and Chris tried to transform with it on sunday.. ligation and transformation failed :-( | ] |
 "
"