Team:INSA-Lyon/Project/Stage3/Results
From 2010.igem.org
| Line 45: | Line 45: | ||
<h3><font color="purple">Curli characterization</font></h3><br><br> | <h3><font color="purple">Curli characterization</font></h3><br><br> | ||
| - | <p>We studied the Curli promoter thanks to the PHL1273 chassis, a mutant OmpR234 bacteria, which contains a reporter gene under the Curli promoter control. This OmpR234 mutation is required for the high expression of the Curli promoter. A modified GFP is used as a reporter gene. This unusual GFP is unstable and has a half-life of 40 min. The fluorescence of the culture is | + | <p>We studied the Curli promoter thanks to the PHL1273 chassis, a mutant OmpR234 bacteria, which contains a reporter gene under the Curli promoter control. This OmpR234 mutation is required for the high expression of the Curli promoter. A modified GFP is used as a reporter gene. This unusual GFP is unstable and has a half-life of 40 min. The fluorescence of the culture is proportional to the quantity of GFP and thus, to the activity of the Curli promoter. |
| - | We chose to study the influence of shaking speed, temperature and osmotic pressure on the activity of this promoter. For all the experiments, we used the same protocol, (the GFP was quantified by fluorescence spectrophotometry, see Protocols). We were three operators to do the same experiment at the same time.</p> | + | We chose to study the influence of shaking speed, temperature and osmotic pressure on the activity of this promoter. For all the experiments, we used the same protocol, (the GFP was quantified by fluorescence spectrophotometry, see <a href="https://2010.igem.org/Team:INSA-Lyon/Project/Notebook/Protocols">protocols</a>). We were three operators to do the same experiment at the same time.</p> |
<p> | <p> | ||
The negative control was a PHL818 chassis, without the reporter plasmid, but which also contains the OmpR234 mutation. The plasmid with Curli promoter and GFP gene was introduced in this chassis in order to create PHL1273 strain. | The negative control was a PHL818 chassis, without the reporter plasmid, but which also contains the OmpR234 mutation. The plasmid with Curli promoter and GFP gene was introduced in this chassis in order to create PHL1273 strain. | ||
| Line 63: | Line 63: | ||
</div> | </div> | ||
| - | <p>First of all, cultures were in a waterbath at 28°C and only the shaking speed was changed. We | + | <p>First of all, cultures were in a waterbath at 28°C and only the shaking speed was changed. We tried 9 different shaking speeds from 50 to 200 rpm.</p> |
<br><br> | <br><br> | ||
| Line 80: | Line 80: | ||
<p> | <p> | ||
| - | The optimal temperature seems to be 28°C. So our system | + | The optimal temperature seems to be 28°C. So our system also presents an optimal response according to the temperature. |
For both series of measurements, GFP fluorescence was very low, while the shaking speed and the temperature were high. Notably, when the temperature is higher than 36°C, the GFP fluorescence is the same as the negative control. | For both series of measurements, GFP fluorescence was very low, while the shaking speed and the temperature were high. Notably, when the temperature is higher than 36°C, the GFP fluorescence is the same as the negative control. | ||
<br> | <br> | ||
| Line 113: | Line 113: | ||
</div> | </div> | ||
| - | <p> We managed a quantification of biofilms formation of a mutant OmpR234 strain (PHL818 chassis, more explained above), versus a wild-type strain (MG1655), at 30°C, in a 24-well polystyrene maden plate. Optimal conditions to observe biofilms is a growth in minimal medium (M63/2), supplemented with a carbon source (glucose 0,2g/L). We modified osmotic pressure | + | <p> We managed a quantification of biofilms formation of a mutant OmpR234 strain (PHL818 chassis, more explained above), versus a wild-type strain (MG1655), at 30°C, in a 24-well polystyrene maden plate. Optimal conditions to observe biofilms is a growth in minimal medium (M63/2), supplemented with a carbon source (glucose 0,2g/L). We modified osmotic pressure the same way as performed with PHL1273 (varying sucrose osmolyte concentration, see above). Then we measured, at 48h culture, biomass in supernatants and also adherent biomass (absorbance at 600nm). Measures were performed in duplicates.<br> |
</p> | </p> | ||
Revision as of 01:07, 28 October 2010
Results
Curli characterization
We studied the Curli promoter thanks to the PHL1273 chassis, a mutant OmpR234 bacteria, which contains a reporter gene under the Curli promoter control. This OmpR234 mutation is required for the high expression of the Curli promoter. A modified GFP is used as a reporter gene. This unusual GFP is unstable and has a half-life of 40 min. The fluorescence of the culture is proportional to the quantity of GFP and thus, to the activity of the Curli promoter. We chose to study the influence of shaking speed, temperature and osmotic pressure on the activity of this promoter. For all the experiments, we used the same protocol, (the GFP was quantified by fluorescence spectrophotometry, see protocols). We were three operators to do the same experiment at the same time.
The negative control was a PHL818 chassis, without the reporter plasmid, but which also contains the OmpR234 mutation. The plasmid with Curli promoter and GFP gene was introduced in this chassis in order to create PHL1273 strain.
Influence of the shaking speed
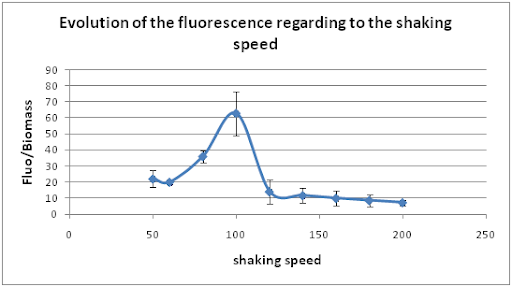
First of all, cultures were in a waterbath at 28°C and only the shaking speed was changed. We tried 9 different shaking speeds from 50 to 200 rpm.
The curve shows that our system presents the nice property to have an optimal shaking speed at 100 rpm when the temperature is 28°C.
Influence of the temperature
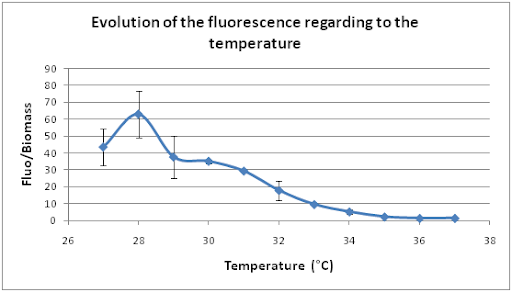
The next series of cultures were in a waterbath with a shaking speed of 100 rpm and we modified the temperature from 27°C to 37°C, degree by degree.
The optimal temperature seems to be 28°C. So our system also presents an optimal response according to the temperature.
For both series of measurements, GFP fluorescence was very low, while the shaking speed and the temperature were high. Notably, when the temperature is higher than 36°C, the GFP fluorescence is the same as the negative control.
Influence of the osmotic pressure
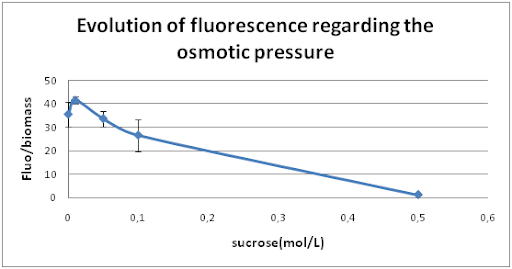
Then, we studied the influence of osmotic pressure on our promoter. We modified the osmotic pressure by changing the sucrose concentration of the culture media. We chose 5 different concentrations in sucrose : 0M - 0,01M - 0,05M - 0,1M – 0,5M. The samples were incubated at 28°C with a shaking speed of 100rpm (previous optimal conditions).
We deduced from this curve that when the osmotic pressure is decreased, the promoter Curli is less active.
In conclusion, the Curli promoter is very responsive at different conditions of culture: shaking speed, temperature and osmotic pressure. Indeed, a gene under the Curli promoter control can be switched ON or OFF by acting on temperature or speed shaking or osmotic pressure. This system is so very versatile and can respond to different environmental signals.
OmpR234 characterization

We managed a quantification of biofilms formation of a mutant OmpR234 strain (PHL818 chassis, more explained above), versus a wild-type strain (MG1655), at 30°C, in a 24-well polystyrene maden plate. Optimal conditions to observe biofilms is a growth in minimal medium (M63/2), supplemented with a carbon source (glucose 0,2g/L). We modified osmotic pressure the same way as performed with PHL1273 (varying sucrose osmolyte concentration, see above). Then we measured, at 48h culture, biomass in supernatants and also adherent biomass (absorbance at 600nm). Measures were performed in duplicates.
As we can see, a decreasing adherence rate
 "
"


