Team:INSA-Lyon/Project/Stage1/Theory
From 2010.igem.org
How to produce a granule?
First a bit of theory, and further our ideas.
Synthesis of PHB
The key enzyme for the PHA biosynthesis is the PHA synthase which polymerizes (R)-3-hydroxyacyl-CoA thioester monomers into polyester.
PhaCAB genes are localized on the first chromosome of bacteria Ralstonia eutropha H16 (old named Alcaligenes eutrophus), (reference: NC_008313):
- The C gene encodes for the 589 amino acids protein, called poly-β-hydroxybutyrate polymerase
(reference: EC 2.3.1.-) - The A gene encodes for the 393 amino acids protein, called acetyl-CoA C-acetyltransferase
(reference: EC 2.3.1.9) - The B gene encodes for the 246 amino acids protein, called acetoacetyl-CoA reductase
(reference: EC 1.1.1.36).
These three proteins seem to be cytoplasmic. They are involved in many metabolic pathways included the bioplastic poly-3-hydroxybutyrate (PHB) synthesis.
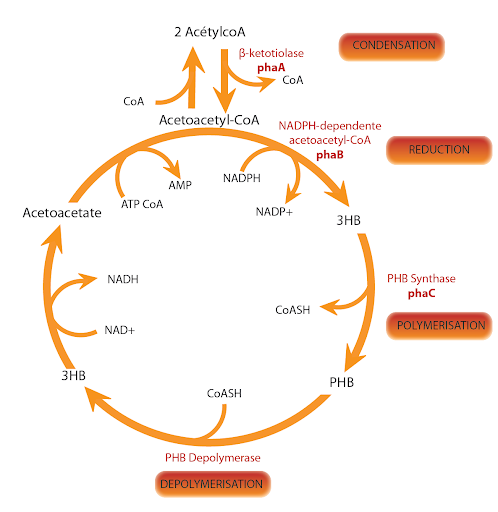
Cycle of the PHB
Biogenesis of granules
There is no consensus about the formation of the PHB granules. Two models have been proposed, and all the experiments realized until May 2009 (Jendrossek 2009) can’t confirm or refute one of them.
But it’s admitted that three proteins play a major part in the biogenesis of the granule : PHB synthase (phaC), PhaPs and PhaR. This two last proteins bind to the granules when those are formed.
The first model is called “micelle model”. It is based on the fact that soluble PHB synthase reacts with its substrate, hydroxyacyl CoA, in the cytoplasm. After the synthesis of the first PHB chain, the others will aggregate to the “primer” by hydrophobic interactions, to make small PHB granules. The enzyme remains in the surface of the granule, and the other proteins of the granule (phasins, PhaR) bind to the growing surface.

Micelle model, from "Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase",Stubbe and Tian, 2003
The second one is called “budding model”. In this one, the PHB synthase is binding the cytoplasmic membrane and the growing PHB chain. The polymer interacts with the cytoplasmic membrane by hydrophobic bonds. We observe the formation of PHB molecules in the cytoplasmic membrane. Later, the granules detach from the membrane, forming structures like “buds”, and the other proteins of the granule (phasins, PhaR) bind to the growing surface.

Budding model, from "Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase",Stubbe and Tian, 2003
Usually, the diameter of the granule range between 100 and 500nm. Various granules are synthesized in the bacteria. The number seems to be controlled by phasins too, but again, no publication can confirm it.The formation of this structure seems to be fast. Indeed, the first granule appears after 10 minutes.
Our ideas
To our knowledge, the production of PHAs has never been observed in wild type E. coli strains. But recombinant strains have been constructed by cloning the genes responsible for the production of PHB, and these molecules were stored into granules (with the diameter usually ranging between 100 and 500 nm and 5-10 granules per cell). These recombinant bacteria are able to accumulate as much as 80% of their dry weight in PHB.
A recombinant MG1655 Escherichia coli has been developed to spontaneously liberate PHB granules (Jung et al., Research in Microbiology, 2005). See our strategy
Evolution
Evolution has been naturally performing synthetic biology for the last thousands years without our knowledge. Evolution combined with mutation and environmental changes have designed and constructed new biological functions and systems not found so far in nature.
We were particularly interested in studying how discrete monofuntional enzymes got organised into one single multifunctional enzyme through evolution. PHB are indeed produced with the help of three distinct enzymes. It would be an interessant goal to increase this lipid production by designing one optimized multifunctional enzyme.
First, we decided to pursue a computer-based approach using databases such that NCBI. We compared FAS I enzyme function for different organisms (mammals and bacteria). We therefore managed to show a high-level of similarity between the domains of the FAS I.
Second, we wanted to analyze these collected data to design a more evolved structure of the operon gene PHA. This is based on the hypothesis that a multi-functional enzyme would increase the yield of lipid production by E.coli.
 "
"


