Team:INSA-Lyon/Project/Stage1/Strategy
From 2010.igem.org
| (38 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
.pili{width: 550px; height: 400px; position:relative; float: center; margin:20px;} | .pili{width: 550px; height: 400px; position:relative; float: center; margin:20px;} | ||
| + | .design{width:300px; height: 100px; position:relative; float:center;} | ||
| + | .design2{width:550px; height: 400px; position:relative; float:center;} | ||
</style> | </style> | ||
| Line 33: | Line 35: | ||
</script> | </script> | ||
| - | + | <a href="top"></a> | |
<div id="corps2"> | <div id="corps2"> | ||
| + | <br> | ||
<h2>Strategy</h2> | <h2>Strategy</h2> | ||
| - | |||
| - | |||
| - | |||
| - | <a href="https://static.igem.org/mediawiki/2010/ | + | <br><br> |
| + | <h3><font color="purple">Complete plasmid synthesis : pILI1</font></h3><br><br> | ||
| + | |||
| + | <p>In 1988, Slater et al. developed the plasmid pSB20. Then the construction of a total DNA bank of Alcaligenes eutrophus permits to identified two restriction sites which surround the three genes and their promoter: KpnI and EcoRI. With the help of a software (Serial Cloner) and the sequence of Ralstonia eutropha (old named Alcaligenes eutrophus), a 5800 pb length sequence had been isolated. It contains the promoter and the phaCAB genes. This sequence had been inserted into a plasmid pUC18. A HindIII site had been added before the promoter and also a unique restriction site XhoI between the promoter and the operon (so we could change the promoter). This is our plasmid pILI1. </p> | ||
| + | |||
| + | <a href="https://static.igem.org/mediawiki/2010/0/0f/Pili.png"> | ||
<div style="text-align:center;"> | <div style="text-align:center;"> | ||
| - | <img class= "pili" src=" | + | <img class= "pili" src="https://static.igem.org/mediawiki/2010/0/0f/Pili.png" alt="Plasmid pILI1 map (Adapted from PlasMapper and BPROM online softwares)" Title="Plasmid pILI1 map (Adapted from PlasMapper and BPROM online softwares)" /> <br/> |
| - | https://static.igem.org/mediawiki/2010/ | + | </a> |
| - | + | <p style="text-align:center; font-size:0.9em;"><em>Plasmid pILI1 map (Adapted from PlasMapper and BPROM online softwares) </em></p> | |
| - | <p style="font-size:0. | + | <br/><br/></div> |
| + | <p>We sent this sequence to GeneCust to be synthesized. The product of this synthesis had been transformed in competent bacteria NM522.</p> | ||
| + | <p>However this plasmid doesn’t match with the iGEM criteria. Indeed some forbidden restriction sites are present in the sequence.</p><br/> | ||
<br/> | <br/> | ||
| - | < | + | <br/> |
| - | < | + | <div style="text-align:center;"> |
| - | < | + | <h3><font color="purple">PhaCAB Operon: two strategies to construct a standard BioBrick</font></h3><br><br> |
| - | < | + | </div> |
| - | <p | + | <p> |
| - | < | + | <h4>Design and PCR</h4><br> |
| - | + | <p> | |
| - | + | We designed a <i>phaC</i> gene in BioBrick standard from the pILI1 sequence: as there were two Pst1 and one Not1 sites inside the gene, we looked for silent mutations in those sites and | |
| + | modified the sequence to make the sites disappear. Then we added the sequence of iGEM prefix at the beginning and the suffix one at the end in order to have the sequence in the standard BioBrick.</p> | ||
<br/> | <br/> | ||
| + | <div style="text-align:center;"> | ||
| + | <img src="http://lh3.ggpht.com/_Uc3bmii-yi0/TMgfmJf-2aI/AAAAAAAAAmY/MmQgbaghPRs/sequenceStrategieStage1.PNG" alt="Sequence" title="Sequence" /> | ||
| + | </div><br/> | ||
| + | <p>(The A from the ATG of the coding sequence completes the Xba1 site)</p> | ||
| + | <br/><br/> | ||
| + | |||
| + | <p>In parallel, we performed a PCR to get <i>phaA</i> and <i>phaB</i> from the pILI1 sequence because no iGEM restriction sites were on the sequence and the sequences of those genes were short enough. We designed them using a software to make them the most stable and the most specific that we could, adding the prefix sequence at the beginning of the forward primer and the suffix sequence at the end of the reverse primer. Then we wanted to ligate the three genes in the original order.</p> | ||
| + | |||
| + | <div style="text-align:center;"> | ||
| + | <img src="http://lh6.ggpht.com/_zZap34AU7o8/TMc9ZuoUPMI/AAAAAAAAABA/agtSWcFEkuc/s640/operon_seq_wiki.png" alt="Strategy with PCR" title="First strategy" /> <br/> | ||
| + | <p style="text-align:center; font-size:0.9em;"><em>First strategy for ligation </em></p> | ||
| + | </div> | ||
<br> | <br> | ||
| + | <br> | ||
| + | <h4>Using iGEM Regestry</h4><br> | ||
| + | <p>We also got the three genes directly from the iGEM registry, and we tried to ligate | ||
| + | them. The BioBricks we used are : <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K156012">BBa_K156012</a> for phaA, <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K156013">BBa_K156013</a> for phaB and <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K156014">BBa_K156014</a> for phaC.</p> | ||
| + | |||
| + | |||
| + | <br /> | ||
| + | <br /> | ||
</div> | </div> | ||
| + | <p style="text-align:center"><a href="#top">Top of Page</a></p> | ||
<div id="corps4" style="border:0px;"> | <div id="corps4" style="border:0px;"> | ||
Latest revision as of 03:55, 28 October 2010
Strategy
Complete plasmid synthesis : pILI1
In 1988, Slater et al. developed the plasmid pSB20. Then the construction of a total DNA bank of Alcaligenes eutrophus permits to identified two restriction sites which surround the three genes and their promoter: KpnI and EcoRI. With the help of a software (Serial Cloner) and the sequence of Ralstonia eutropha (old named Alcaligenes eutrophus), a 5800 pb length sequence had been isolated. It contains the promoter and the phaCAB genes. This sequence had been inserted into a plasmid pUC18. A HindIII site had been added before the promoter and also a unique restriction site XhoI between the promoter and the operon (so we could change the promoter). This is our plasmid pILI1.

Plasmid pILI1 map (Adapted from PlasMapper and BPROM online softwares)
We sent this sequence to GeneCust to be synthesized. The product of this synthesis had been transformed in competent bacteria NM522.
However this plasmid doesn’t match with the iGEM criteria. Indeed some forbidden restriction sites are present in the sequence.
PhaCAB Operon: two strategies to construct a standard BioBrick
Design and PCR
We designed a phaC gene in BioBrick standard from the pILI1 sequence: as there were two Pst1 and one Not1 sites inside the gene, we looked for silent mutations in those sites and modified the sequence to make the sites disappear. Then we added the sequence of iGEM prefix at the beginning and the suffix one at the end in order to have the sequence in the standard BioBrick.
(The A from the ATG of the coding sequence completes the Xba1 site)
In parallel, we performed a PCR to get phaA and phaB from the pILI1 sequence because no iGEM restriction sites were on the sequence and the sequences of those genes were short enough. We designed them using a software to make them the most stable and the most specific that we could, adding the prefix sequence at the beginning of the forward primer and the suffix sequence at the end of the reverse primer. Then we wanted to ligate the three genes in the original order.
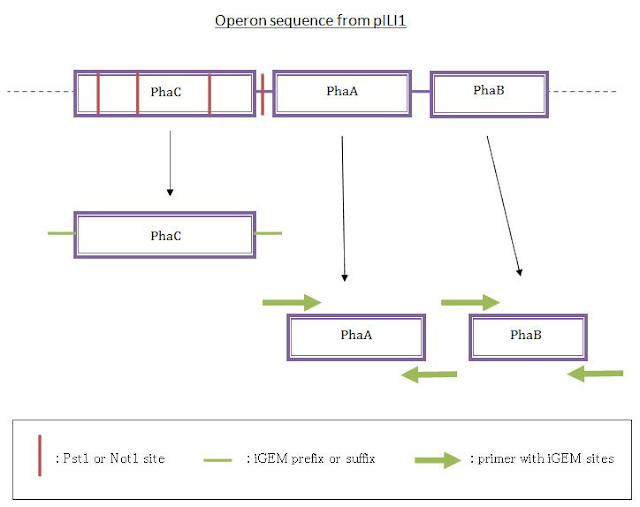
First strategy for ligation
Using iGEM Regestry
We also got the three genes directly from the iGEM registry, and we tried to ligate them. The BioBricks we used are : BBa_K156012 for phaA, BBa_K156013 for phaB and BBa_K156014 for phaC.
 "
"


