Team:INSA-Lyon/Project/Stage1/Results
From 2010.igem.org
| (12 intermediate revisions not shown) | |||
| Line 34: | Line 34: | ||
<div id="corps2"> | <div id="corps2"> | ||
| - | |||
| - | |||
<br> | <br> | ||
| - | < | + | <h2>Results</h2> |
| + | <br><br> | ||
| + | <h3 style="color:purple;"><b>The pILI1 plasmid</b></h3><br/> | ||
| - | <p>The transformed bacteria had been spread on LB | + | <p>The transformed bacteria (containing the pILI1 plasmid) had been spread on LB agar plates, with or without 7% of glucose and RedNile dye. As you can see on the photos below, the bacteria appear pink colored, directly on plates supplemented with glucose. However, the control without glucose doesn't reveal a pink coloration.</p><br/> |
| - | <p>With a liquid culture on LB complemented with glucose (7%) and RedNile, we observed the fluorescence by microscopy.</p><br/> | + | <div style="text-align:center;"> |
| + | <img src="http://lh4.ggpht.com/_Uc3bmii-yi0/TMf__pnKyLI/AAAAAAAAAl4/KBXck8LZerk/P1050468%20%282%29.JPG" /> | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | <p style="font-size:0.9em; text-indent:0px; text-align:center;"><em>LB agar plate with pILI1, with or without glucose 7% and RedNile dye.</em><br/><br/><br/></p> | ||
| + | </div><br/> | ||
| + | |||
| + | <p>With a liquid culture on LB complemented with glucose (7%) and RedNile dye, we observed the fluorescence by microscopy.</p><br/> | ||
<div style="text-align:center;"> | <div style="text-align:center;"> | ||
| - | <img src="http:// | + | <img src="http://lh6.ggpht.com/_Uc3bmii-yi0/TMgFqr4biTI/AAAAAAAAAmQ/Lp_33O0Td8c/Granule1.jpg" /> |
| - | <img src="http:// | + | <img src="http://lh5.ggpht.com/_Uc3bmii-yi0/TMgEslmyE_I/AAAAAAAAAmE/ODeTOjHAb4g/Granule2.jpg" /> <br/> |
| - | <p style="font-size:0. | + | <p style="font-size:0.9em; text-indent:0px; text-align:center;"><em>Observation of fluorescent granules by microscopy. </em><br/><br/><br/></p> |
</div> | </div> | ||
| - | < | + | <br> |
| - | <br/><p>We | + | <h3 style="color:purple;">The separated genes</h3> |
| + | <br/><h4>Design and PCR</h4><br> | ||
| + | <p>We cloned the <span style="font-style:italic">phaC</span> gene from the synthesized plasmid (Mr Gene), into the pSB1C3 backbone. After sequencing (EurofinDNA) checked, we sent the cloning product to the registry: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K342001">BBa_K342001</a><br/><br/></p> | ||
| + | <br> | ||
| + | <h4>Using iGEM Registry</h4><br> | ||
| + | <p>When we tried to check the final ligation, we realized that we weren't able to cut either the new operon either the phaC gene with the Xba1 enzyme : when cutting by Xba1 and Pst1 we obtained a single band corresponding to the size of our whole plasmid. When cutting by Pst1 only, we obtained the same result whereas when we tried to cut by Xba1 we got several bands like for a circular plasmid. | ||
| + | We then tried to cut the phaC gene with Xba1 and we aslo obtained the same pattern as the one of a circular plasmid. <br/> | ||
| + | With more time we could have tried to design a primer with the iGEM restriction sites and see if we were able to cut the new sequence.</p> | ||
| + | <br> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | <p style="text-align:center;"><a href="#top">Top of Page</a></p> | ||
<div id="corps4" style="border:0px;"> | <div id="corps4" style="border:0px;"> | ||
Latest revision as of 21:32, 27 October 2010
Results
The pILI1 plasmid
The transformed bacteria (containing the pILI1 plasmid) had been spread on LB agar plates, with or without 7% of glucose and RedNile dye. As you can see on the photos below, the bacteria appear pink colored, directly on plates supplemented with glucose. However, the control without glucose doesn't reveal a pink coloration.
LB agar plate with pILI1, with or without glucose 7% and RedNile dye.
With a liquid culture on LB complemented with glucose (7%) and RedNile dye, we observed the fluorescence by microscopy.
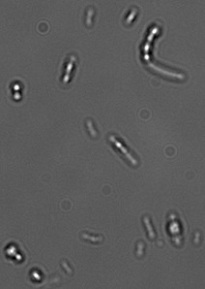
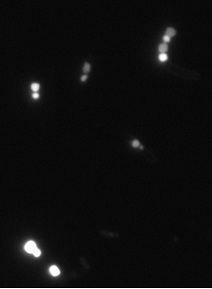
Observation of fluorescent granules by microscopy.
The separated genes
Design and PCR
We cloned the phaC gene from the synthesized plasmid (Mr Gene), into the pSB1C3 backbone. After sequencing (EurofinDNA) checked, we sent the cloning product to the registry: BBa_K342001
Using iGEM Registry
When we tried to check the final ligation, we realized that we weren't able to cut either the new operon either the phaC gene with the Xba1 enzyme : when cutting by Xba1 and Pst1 we obtained a single band corresponding to the size of our whole plasmid. When cutting by Pst1 only, we obtained the same result whereas when we tried to cut by Xba1 we got several bands like for a circular plasmid.
We then tried to cut the phaC gene with Xba1 and we aslo obtained the same pattern as the one of a circular plasmid.
With more time we could have tried to design a primer with the iGEM restriction sites and see if we were able to cut the new sequence.
 "
"


