Team:HokkaidoU Japan/Notebook/September3
From 2010.igem.org
(Difference between revisions)
(New page: {{Template:HokkaidoU_Japan}}) |
|||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:HokkaidoU_Japan}} | + | {{Template:HokkaidoU_Japan}}<div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/September2|September 2]]</div>[[Team:HokkaidoU_Japan/Notebook|Notebook]]<div class="next">[[Team:HokkaidoU_Japan/Notebook/September6|September 6]]</div></div> |
| + | |||
| + | =Results of yesterdays transformation= | ||
| + | * pSB1C3 uterly failed to produce colonies | ||
| + | * pUC119 produced 20 colonies | ||
| + | * colonies that should been red because of RFP insert wasn't, so there is possibility that insert wasn't there | ||
| + | |||
| + | =Colony PCR on yesterdays E.coli= | ||
| + | * Colony PCR was done according to protocol | ||
| + | * This day we did 20 samples | ||
| + | |||
| + | ===Electrophoresis=== | ||
| + | |||
| + | =Concentration check of parts used for transformation yesterday= | ||
| + | |||
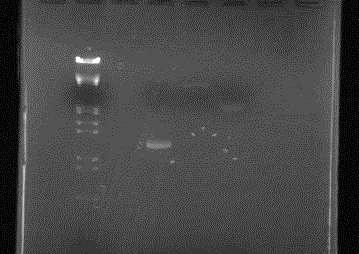
| + | [[Image:HokkaidoU_Japan_20100903a.jpg|200px|right|thumb|Electrophoresis of parts before ligation]] | ||
| + | |||
| + | Confirmed that DNA solution concentration of parts for Ligation was as anticipated (good?) | ||
| + | |||
| + | {|class="protocol" | ||
| + | |- | ||
| + | |'''Lane''' | ||
| + | |'''DNA''' | ||
| + | |- | ||
| + | |1 | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |[https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png λ/Hind III, EcoR I] | ||
| + | |- | ||
| + | |3 | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |RFP | ||
| + | |- | ||
| + | |5 | ||
| + | |pUC119 | ||
| + | |- | ||
| + | |6 | ||
| + | |pSB1C3 | ||
| + | |} | ||
| + | |||
| + | =[[Team:HokkaidoU_Japan/Protocols|PCR]] of [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]]= | ||
| + | |||
| + | [[Team:HokkaidoU_Japan/Protocols|Transformation]] of [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] part which is originaly on pSB1C3 was succesful<br> | ||
| + | Thinking that linearized vector might gone bad we PCRed pSB1C3 from [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] | ||
| + | |||
| + | {|style="text-align:center;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |[[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] | ||
| + | |1 | ||
| + | |- | ||
| + | |DW | ||
| + | |33 | ||
| + | |- | ||
| + | |10x Buffer | ||
| + | |5 | ||
| + | |- | ||
| + | |2 M 4dNTPs | ||
| + | |5 | ||
| + | |- | ||
| + | |25 mM MgSO<sub>4</sub> | ||
| + | |3 | ||
| + | |- | ||
| + | |Suffix-F | ||
| + | |1 | ||
| + | |- | ||
| + | |Prefix-R | ||
| + | |1 | ||
| + | |- | ||
| + | |KOD | ||
| + | |1 | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''50 uL''' | ||
| + | |} | ||
| + | |||
| + | * Extension was for120 sec | ||
| + | * Purified via Microcon YM-10 | ||
| + | * Final volume was 43 uL | ||
Latest revision as of 08:07, 27 October 2010
Results of yesterdays transformation
- pSB1C3 uterly failed to produce colonies
- pUC119 produced 20 colonies
- colonies that should been red because of RFP insert wasn't, so there is possibility that insert wasn't there
Colony PCR on yesterdays E.coli
- Colony PCR was done according to protocol
- This day we did 20 samples
Electrophoresis
Concentration check of parts used for transformation yesterday
Confirmed that DNA solution concentration of parts for Ligation was as anticipated (good?)
| Lane | DNA |
| 1 | |
| 2 | λ/Hind III, EcoR I |
| 3 | |
| 4 | RFP |
| 5 | pUC119 |
| 6 | pSB1C3 |
PCR of 1-3A
Transformation of 1-3A part which is originaly on pSB1C3 was succesful
Thinking that linearized vector might gone bad we PCRed pSB1C3 from 1-3A
| Reagent | Amount |
|---|---|
| 1-3A | 1 |
| DW | 33 |
| 10x Buffer | 5 |
| 2 M 4dNTPs | 5 |
| 25 mM MgSO4 | 3 |
| Suffix-F | 1 |
| Prefix-R | 1 |
| KOD | 1 |
| Total | 50 uL |
- Extension was for120 sec
- Purified via Microcon YM-10
- Final volume was 43 uL
 "
"