Team:HokkaidoU Japan/Notebook/September2
From 2010.igem.org
(Difference between revisions)
(New page: {{Template:HokkaidoU_Japan}}) |
|||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:HokkaidoU_Japan}} | + | {{Template:HokkaidoU_Japan}}<div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/September1|September 1]]</div>[[Team:HokkaidoU_Japan/Notebook|Notebook]]<div class="next">[[Team:HokkaidoU_Japan/Notebook/September3|September 3]]</div></div> |
| + | |||
| + | =[[Team:HokkaidoU_Japan/Protocols|Transformation]] of pUC119, RFP, pSB1C3 and RFP= | ||
| + | |||
| + | ==Parts information== | ||
| + | {|style="text-align:center; class="protocol" | ||
| + | |- | ||
| + | !DNA | ||
| + | !Concentration | ||
| + | !Length | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP ([[Team:HokkaidoU_Japan/Parts#BioBricks|1-5A]]) | ||
| + | |80 ng/uL | ||
| + | |1000 bp | ||
| + | |26 uL | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |20 ng/uL | ||
| + | |2000 bp | ||
| + | |35 uL | ||
| + | |- | ||
| + | |pUC119(pre cut) | ||
| + | |10 ng/uL | ||
| + | |3000 bp | ||
| + | |28 uL | ||
| + | |} | ||
| + | |||
| + | ==Digestion== | ||
| + | |||
| + | {|style="text-align:center; float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP ([[Team:HokkaidoU_Japan/Parts#BioBricks|1-5A]]) | ||
| + | |26 uL | ||
| + | |- | ||
| + | |DW | ||
| + | |9 uL | ||
| + | |- | ||
| + | |0.1% BSA | ||
| + | |5 uL | ||
| + | |- | ||
| + | |10x M Buffer | ||
| + | |5 uL | ||
| + | |- | ||
| + | |EcoR I | ||
| + | |2 uL | ||
| + | |- | ||
| + | |Pst I | ||
| + | |3 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''50 uL''' | ||
| + | |} | ||
| + | |||
| + | {|style="text-align:center; float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |33 uL | ||
| + | |- | ||
| + | |DW | ||
| + | |2 uL | ||
| + | |- | ||
| + | |0.1% BSA | ||
| + | |5 uL | ||
| + | |- | ||
| + | |10x M Buffer | ||
| + | |5 uL | ||
| + | |- | ||
| + | |EcoR I | ||
| + | |2 uL | ||
| + | |- | ||
| + | |Pst I | ||
| + | |3 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''50 uL''' | ||
| + | |} | ||
| + | <div style="clear:both;"></div> | ||
| + | →Incubated at 37C for 60 min<br> | ||
| + | →Adde 10 uL 6x Sample buffer and electrophoresed | ||
| + | |||
| + | ==Electrophoresis and [[Team:HokkaidoU_Japan/Protocols|gel extraction]]== | ||
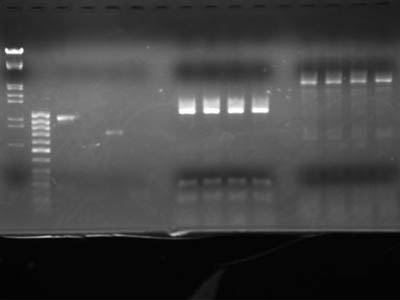
| + | To check concentration lanes 2 through 5 contained 1uL of DNA solution | ||
| + | [[Image:HokkaidoU Japan 20100902a.jpg|200px|right|thumb|Electrophoresis of digestion product]] | ||
| + | {|class="protocol" | ||
| + | |'''Lane''' | ||
| + | |'''DNA''' | ||
| + | |- | ||
| + | |1 | ||
| + | |[https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png λ/''Hin''d III, EcoR I] | ||
| + | |- | ||
| + | |2 | ||
| + | |50 base ladder | ||
| + | |- | ||
| + | |3 | ||
| + | |Heat shock promotor | ||
| + | |- | ||
| + | |4 | ||
| + | |RBS | ||
| + | |- | ||
| + | |5 | ||
| + | |RFP protein coding | ||
| + | |- | ||
| + | |6 | ||
| + | |double Terminator | ||
| + | |- | ||
| + | |7 | ||
| + | |blank | ||
| + | |- | ||
| + | |8 | ||
| + | |style="border-left:1px solid #000;" rowspan="4"|RFP reporter ([[Team:HokkaidoU_Japan/Parts#BioBricks|1-5A]]) 15 uL each | ||
| + | |- | ||
| + | |9 | ||
| + | |- | ||
| + | |10 | ||
| + | |- | ||
| + | |11 | ||
| + | |- | ||
| + | |12 | ||
| + | |blank | ||
| + | |- | ||
| + | |13 | ||
| + | |rowspan="4" style="border-left:1px solid #000;"|pSB1C3 15 uL each | ||
| + | |- | ||
| + | |14 | ||
| + | |- | ||
| + | |15 | ||
| + | |- | ||
| + | |16 | ||
| + | |- | ||
| + | |17 | ||
| + | |blank | ||
| + | |} | ||
| + | Bands were exised (210 mg and 220 mg acordingly) | ||
| + | Final volume was made to be 20 uL, with disregard to promega's protocol<br> | ||
| + | At this point concentrations were: RFP 104 ng/uL,pSB1C3 33 ng/uL. | ||
| + | |||
| + | ==Ligation== | ||
| + | {|style="text-align:center;float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP | ||
| + | |1.5 uL | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |1.5 uL | ||
| + | |- | ||
| + | |ligation Kit I | ||
| + | |3 uL | ||
| + | |- | ||
| + | |T4 ligase | ||
| + | |0.5 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''6.5 uL''' | ||
| + | |} | ||
| + | |||
| + | {|style="text-align:center; float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP | ||
| + | |1.5 uL | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |1.5 uL | ||
| + | |- | ||
| + | |Mighty Mix | ||
| + | |3 uL | ||
| + | |- | ||
| + | |T4 ligase | ||
| + | |0.5 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''6.5 uL''' | ||
| + | |} | ||
| + | |||
| + | {|style="text-align:center; float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP | ||
| + | |0.8 uL | ||
| + | |- | ||
| + | |pUC119 | ||
| + | |4 uL | ||
| + | |- | ||
| + | |ligation Kit I | ||
| + | |4.8 uL | ||
| + | |- | ||
| + | |T4 ligase | ||
| + | |0.5 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''10.5 uL''' | ||
| + | |} | ||
| + | |||
| + | {|style="text-align:center; float: left;" class="protocol" | ||
| + | !Reagent | ||
| + | !Amount | ||
| + | |- | ||
| + | |RFP | ||
| + | |0.8 uL | ||
| + | |- | ||
| + | |pUC119 | ||
| + | |4 uL | ||
| + | |- | ||
| + | |Mighty Mix | ||
| + | |4.8 uL | ||
| + | |- | ||
| + | |T4 ligase | ||
| + | |0.5 uL | ||
| + | |- | ||
| + | |style="border-top:1px solid #996;"|'''Total''' | ||
| + | |style="border-top:1px solid #996;"|'''10.5 uL''' | ||
| + | |} | ||
| + | <div style="clear:both;"></div> | ||
| + | →Incubated 16C for min<br> | ||
| + | →[[Team:HokkaidoU_Japan/Protocols|Transformation]] | ||
Latest revision as of 08:02, 27 October 2010
Transformation of pUC119, RFP, pSB1C3 and RFP
Parts information
| DNA | Concentration | Length | Amount |
|---|---|---|---|
| RFP (1-5A) | 80 ng/uL | 1000 bp | 26 uL |
| pSB1C3 | 20 ng/uL | 2000 bp | 35 uL |
| pUC119(pre cut) | 10 ng/uL | 3000 bp | 28 uL |
Digestion
| Reagent | Amount |
|---|---|
| RFP (1-5A) | 26 uL |
| DW | 9 uL |
| 0.1% BSA | 5 uL |
| 10x M Buffer | 5 uL |
| EcoR I | 2 uL |
| Pst I | 3 uL |
| Total | 50 uL |
| Reagent | Amount |
|---|---|
| pSB1C3 | 33 uL |
| DW | 2 uL |
| 0.1% BSA | 5 uL |
| 10x M Buffer | 5 uL |
| EcoR I | 2 uL |
| Pst I | 3 uL |
| Total | 50 uL |
→Incubated at 37C for 60 min
→Adde 10 uL 6x Sample buffer and electrophoresed
Electrophoresis and gel extraction
To check concentration lanes 2 through 5 contained 1uL of DNA solution
| Lane | DNA |
| 1 | λ/Hind III, EcoR I |
| 2 | 50 base ladder |
| 3 | Heat shock promotor |
| 4 | RBS |
| 5 | RFP protein coding |
| 6 | double Terminator |
| 7 | blank |
| 8 | RFP reporter (1-5A) 15 uL each |
| 9 | |
| 10 | |
| 11 | |
| 12 | blank |
| 13 | pSB1C3 15 uL each |
| 14 | |
| 15 | |
| 16 | |
| 17 | blank |
Bands were exised (210 mg and 220 mg acordingly)
Final volume was made to be 20 uL, with disregard to promega's protocol
At this point concentrations were: RFP 104 ng/uL,pSB1C3 33 ng/uL.
Ligation
| Reagent | Amount |
|---|---|
| RFP | 1.5 uL |
| pSB1C3 | 1.5 uL |
| ligation Kit I | 3 uL |
| T4 ligase | 0.5 uL |
| Total | 6.5 uL |
| Reagent | Amount |
|---|---|
| RFP | 1.5 uL |
| pSB1C3 | 1.5 uL |
| Mighty Mix | 3 uL |
| T4 ligase | 0.5 uL |
| Total | 6.5 uL |
| Reagent | Amount |
|---|---|
| RFP | 0.8 uL |
| pUC119 | 4 uL |
| ligation Kit I | 4.8 uL |
| T4 ligase | 0.5 uL |
| Total | 10.5 uL |
| Reagent | Amount |
|---|---|
| RFP | 0.8 uL |
| pUC119 | 4 uL |
| Mighty Mix | 4.8 uL |
| T4 ligase | 0.5 uL |
| Total | 10.5 uL |
→Incubated 16C for min
 "
"