Team:HokkaidoU Japan/Notebook/August25
From 2010.igem.org
(Difference between revisions)
(→Estimation of 1-3A Consentration) |
|||
| (5 intermediate revisions not shown) | |||
| Line 19: | Line 19: | ||
|yes | |yes | ||
|- | |- | ||
| - | |1-3A 34 | + | |[[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] 34 |
|yes | |yes | ||
|- | |- | ||
| - | |1-3A 12 | + | |[[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] 12 |
|yes | |yes | ||
|} | |} | ||
<hr width="50%"> | <hr width="50%"> | ||
| - | * 1-3A had colonies in lated on mediums containing 34 ug/uL of chloramphenicol | + | * [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] had colonies in lated on mediums containing 34 ug/uL of chloramphenicol |
** So the medium and antibiotic consentration seems ok, | ** So the medium and antibiotic consentration seems ok, | ||
* Medium on which M 12 was plated was inoculated with chloramphenicol after solidification so there were some irregularities and colonies on the fringe | * Medium on which M 12 was plated was inoculated with chloramphenicol after solidification so there were some irregularities and colonies on the fringe | ||
| - | * Colonies of 1-3A and 12 were obviously different | + | * Colonies of [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] and 12 were obviously different |
→ So we suspected issues in Ligation and DNA concentration bused for transformation | → So we suspected issues in Ligation and DNA concentration bused for transformation | ||
| Line 37: | Line 37: | ||
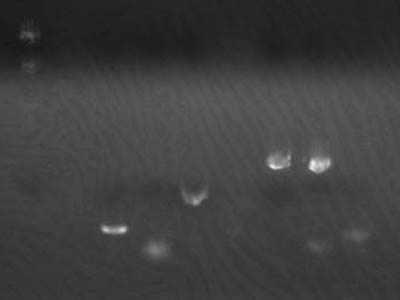
[[Image:HokkaidoU Japan 20100825a.jpg|200px|right|thumb|Electrophoresis of 1uL of 1-3A DNA]] | [[Image:HokkaidoU Japan 20100825a.jpg|200px|right|thumb|Electrophoresis of 1uL of 1-3A DNA]] | ||
| - | *Added 1 uL of 1-3A into 30 uL of competent cells | + | *Added 1 uL of [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]] into 30 uL of competent cells |
*Transformation was successful | *Transformation was successful | ||
| - | *Electrophoresed 1 uL of 1- | + | *Electrophoresed 1 uL of [[Team:HokkaidoU_Japan/Parts#BioBricks|1-3A]],calculated amount of transformed DNA |
→Estimated concentration 2 ng/uL | →Estimated concentration 2 ng/uL | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | = | + | =Ethanol precipitation of pSB1C3 and Ligation= |
| - | === | + | |
| - | # | + | ===Ethanol precipitation=== |
| - | # 100% | + | |
| - | # | + | # Added 8.2 uL of sodium acetate [3 M] |
| - | # | + | # Added 205 uL of Ethanol [100%] and mixed |
| - | # | + | # Centrifuge at 15000rpm, 4C for 10 min |
| - | # | + | # Discarded supernatant and added 500 uL of Ethanol [70%], mixed |
| - | # | + | # Centrifuge at 15000rpm, 4C for 5 min |
| + | # Discarded supernatant | ||
| + | # Dried via vacuum desiccator | ||
| + | # Added 8.2 uL of ADW | ||
| + | * Concentration increased 10 fold | ||
===Ligation=== | ===Ligation=== | ||
| - | # 8.2 uL DNA | + | # Mixed 8.2 uL of DNA solution with the same volume of Ligation Solution |
| - | # T4 ligase | + | # Added extra 1 uL of T4 ligase |
| - | # | + | # Incubated at 16C for 30 min |
| + | Tomorrow will do aditional Electrophoresis gel extraction and transformation | ||
| + | |||
| + | =Digestion of PCR products amplified using digestion visualization primers= | ||
| + | |||
| + | |||
| + | [[Image:HokkaidoU Japan 20100825c.JPG |200px|right|thumb|Electrophoresis after digestion, differences in length is visible]] | ||
| + | |||
| + | Calculated the concentration of DNA amplified yesterday | ||
| - | |||
| - | |||
| - | |||
{| | {| | ||
| Line 77: | Line 86: | ||
|} | |} | ||
| - | ===Digestion | + | ===Digestion Reaction=== |
{|style="text-align:center;" class="protocol" | {|style="text-align:center;" class="protocol" | ||
!Reagent | !Reagent | ||
| Line 104: | Line 113: | ||
|} | |} | ||
| - | + | →Incubated at 37C for 60 min | |
| - | + | →Confirmed how awesome new primers are | |
Latest revision as of 07:50, 27 October 2010
Transformation Results
Colony Check
| Plate | Colonies |
| M 34 | no |
| M 12 | yes |
| I 34 | no |
| I 34 | yes |
| 1-3A 34 | yes |
| 1-3A 12 | yes |
- 1-3A had colonies in lated on mediums containing 34 ug/uL of chloramphenicol
- So the medium and antibiotic consentration seems ok,
- Medium on which M 12 was plated was inoculated with chloramphenicol after solidification so there were some irregularities and colonies on the fringe
- Colonies of 1-3A and 12 were obviously different
→ So we suspected issues in Ligation and DNA concentration bused for transformation
Estimation of 1-3A Consentration
- Added 1 uL of 1-3A into 30 uL of competent cells
- Transformation was successful
- Electrophoresed 1 uL of 1-3A,calculated amount of transformed DNA
→Estimated concentration 2 ng/uL
Ethanol precipitation of pSB1C3 and Ligation
Ethanol precipitation
- Added 8.2 uL of sodium acetate [3 M]
- Added 205 uL of Ethanol [100%] and mixed
- Centrifuge at 15000rpm, 4C for 10 min
- Discarded supernatant and added 500 uL of Ethanol [70%], mixed
- Centrifuge at 15000rpm, 4C for 5 min
- Discarded supernatant
- Dried via vacuum desiccator
- Added 8.2 uL of ADW
- Concentration increased 10 fold
Ligation
- Mixed 8.2 uL of DNA solution with the same volume of Ligation Solution
- Added extra 1 uL of T4 ligase
- Incubated at 16C for 30 min
Tomorrow will do aditional Electrophoresis gel extraction and transformation
Digestion of PCR products amplified using digestion visualization primers
Calculated the concentration of DNA amplified yesterday
| RBS | 27 ng/uL | 312 bp |
| GFP | 54 ng/uL | 1020 bp |
| dT | 49 ng/uL | 429 bp |
Digestion Reaction
| Reagent | Amount |
|---|---|
| 10x M buffer | 1 uL |
| DW | 4.5 |
| DNA | 2.5 |
| BSA | 1 |
| EcoR I | 0.5 |
| Pst I | 0.5 |
| Total | 10 uL |
→Incubated at 37C for 60 min
→Confirmed how awesome new primers are "
"