Team:Heidelberg/Notebook/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Laura Nadine (Talk | contribs) |
||
| Line 731: | Line 731: | ||
<br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| + | = 01/10/2010 = | ||
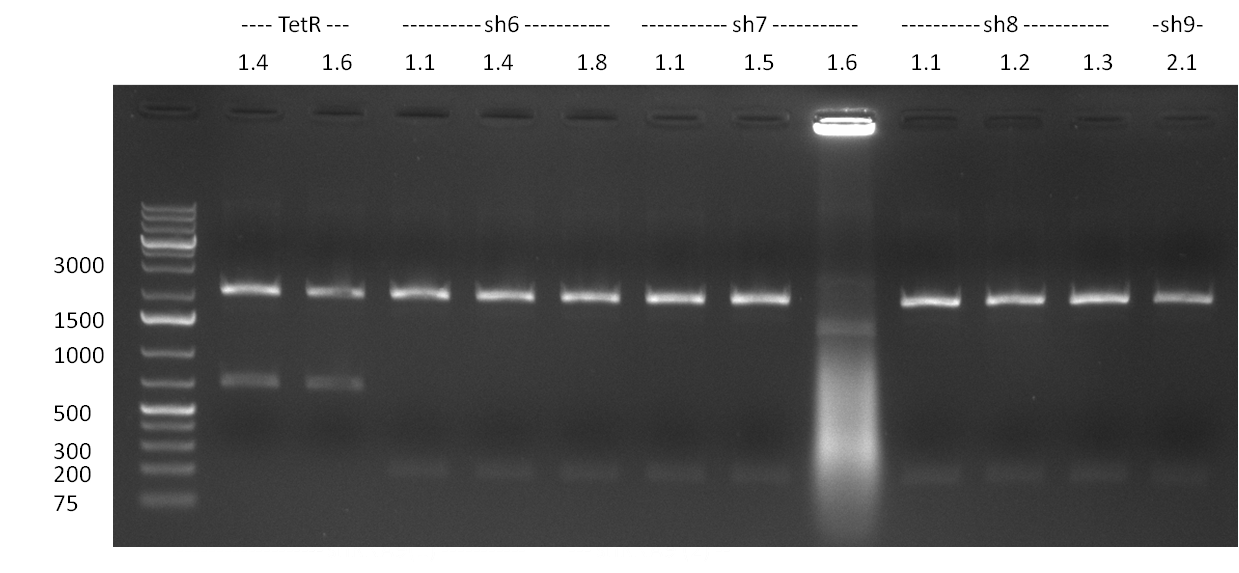
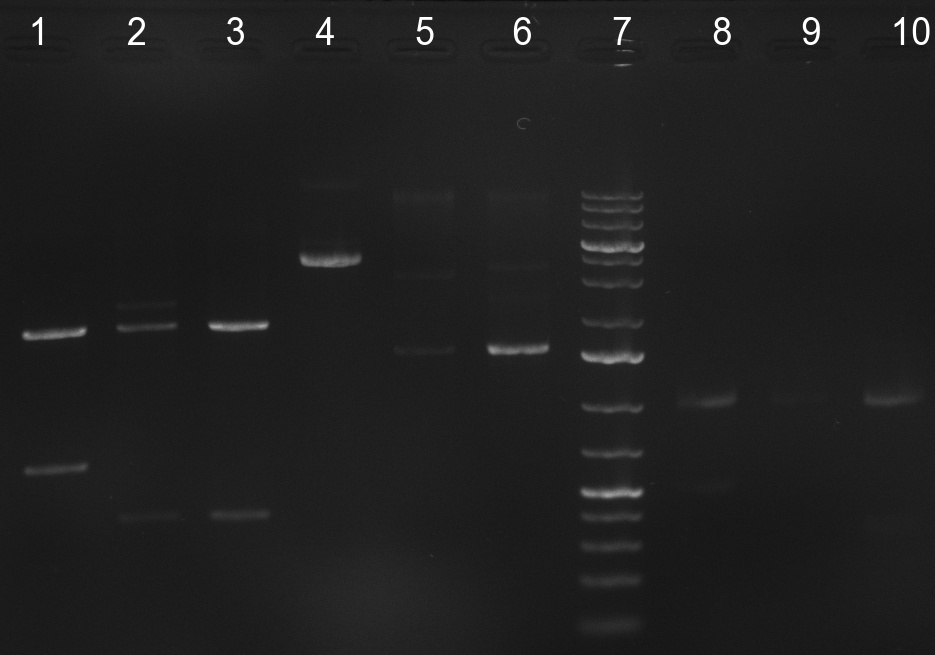
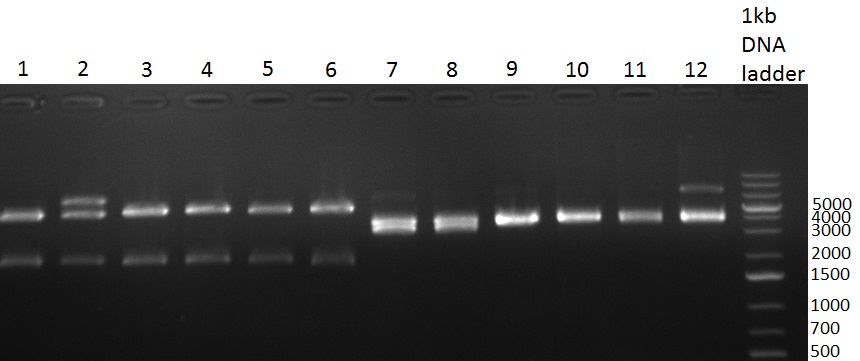
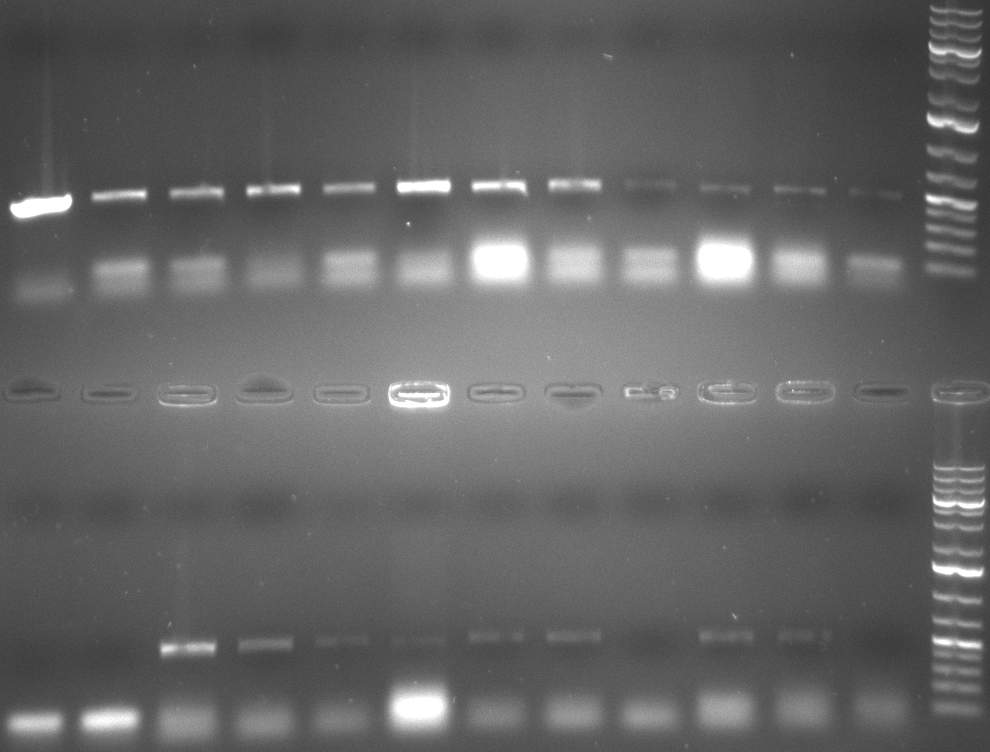
| + | [[Image:20101001_tetR_verdauEPanno.jpg|thumb|300px|right|Gel 101001-1. Digestion of miniprep no. 8, 9, 14 and 16 with EcoRI and PstI. Odd lanes are undigested controls, even lanes are digested minis]] | ||
| + | |||
| + | repressor construct: | ||
| + | * colony 8, 9, 14 and 16 were chosen for miniprep. They consist of the complete TetR construct in pBS 1C3 | ||
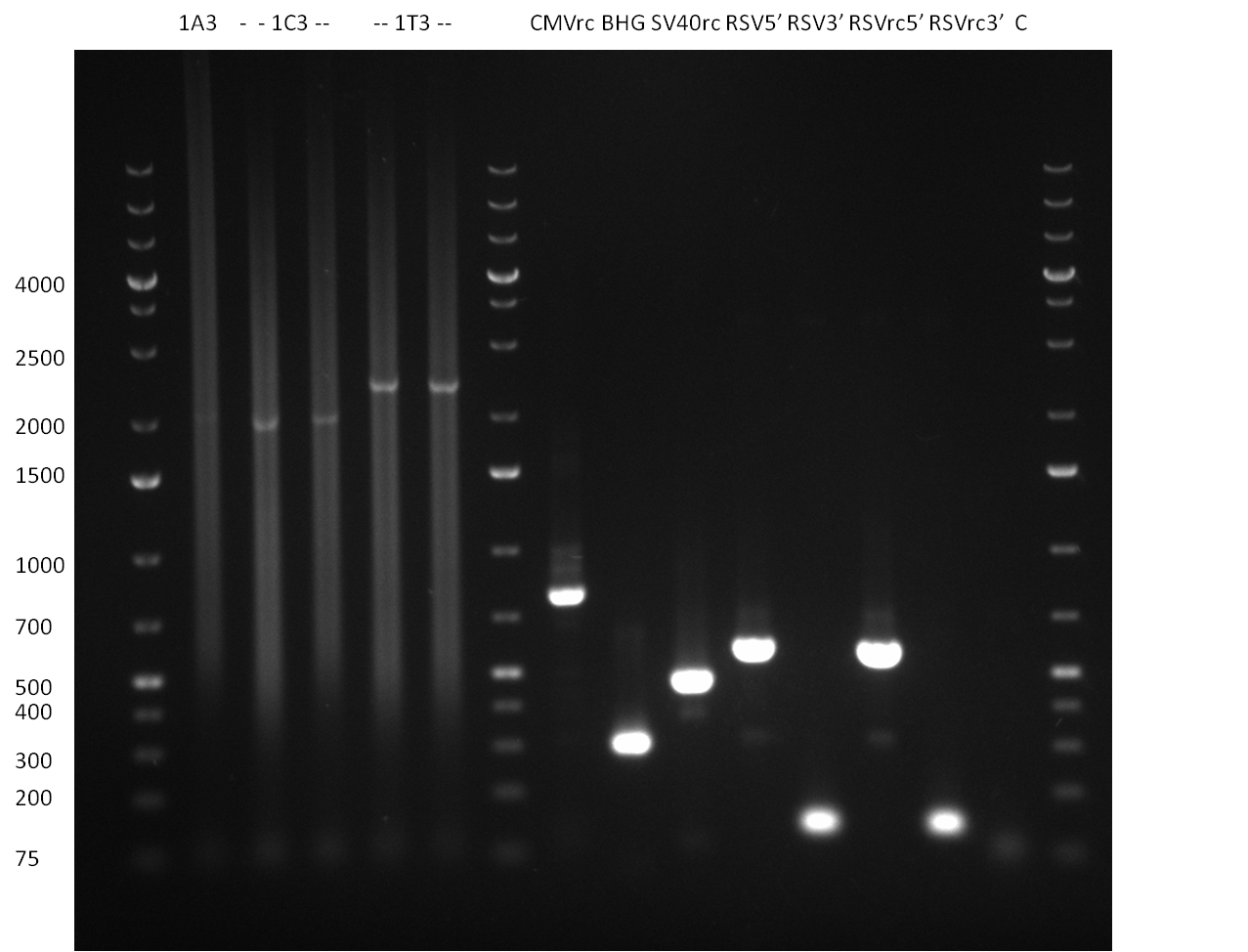
| + | * digestion of the minis with EcoRI and PstI for cloning into {{Part|pSB1A3}}, SAP of {{Part|pSB1A3}}, digestion was tested on gel and proven positive (see Gel 101001-1) | ||
| + | * nucleotide removal, then ligation of TetR8 and TetR14 equimolar with SAPed backbone, TetR9 and TetR16 2:1 with not-SAPed backbone, transformation with TOP10 cells (10µl on 50µl ''E. coli'') | ||
| + | <br /> | ||
| + | <br /> | ||
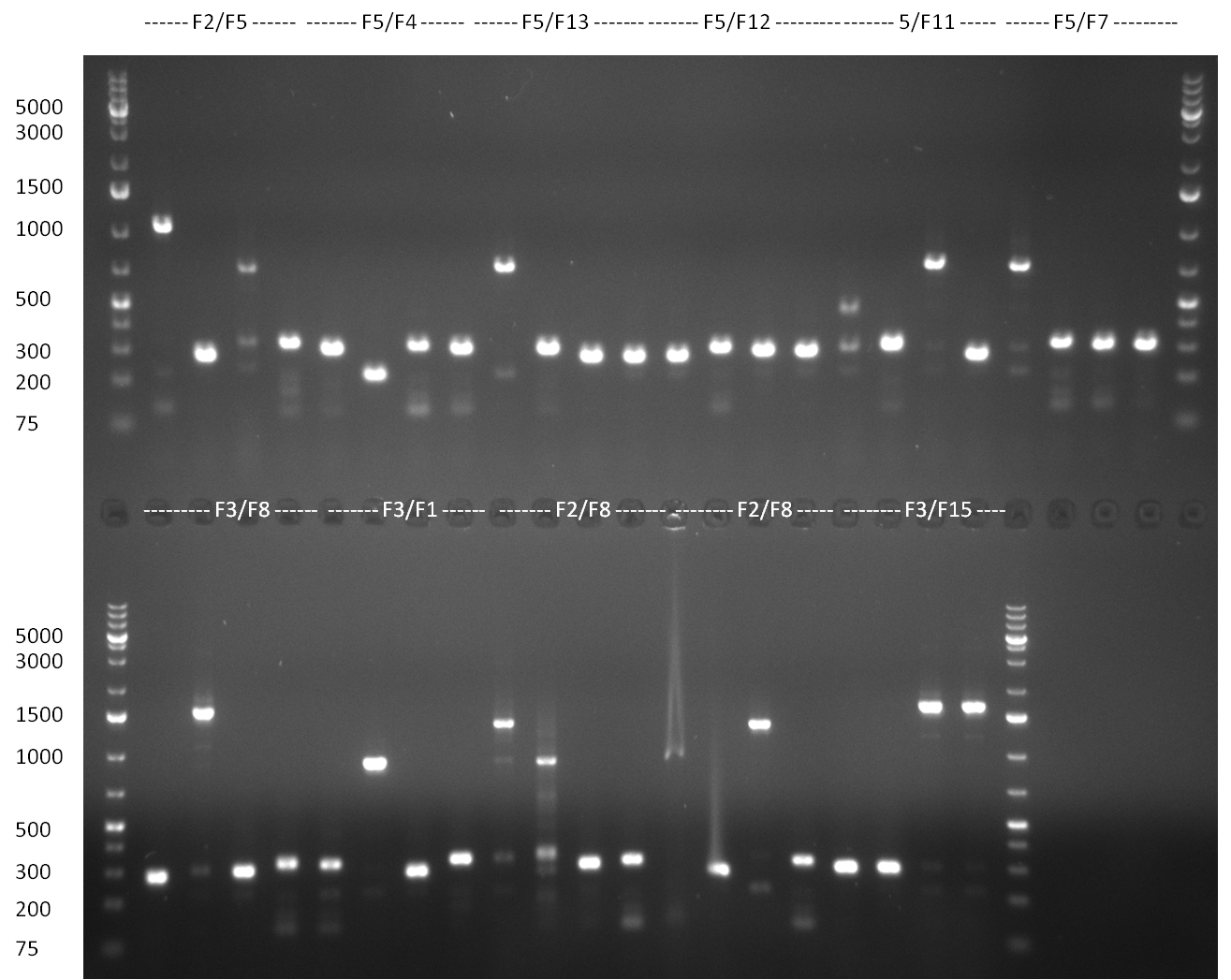
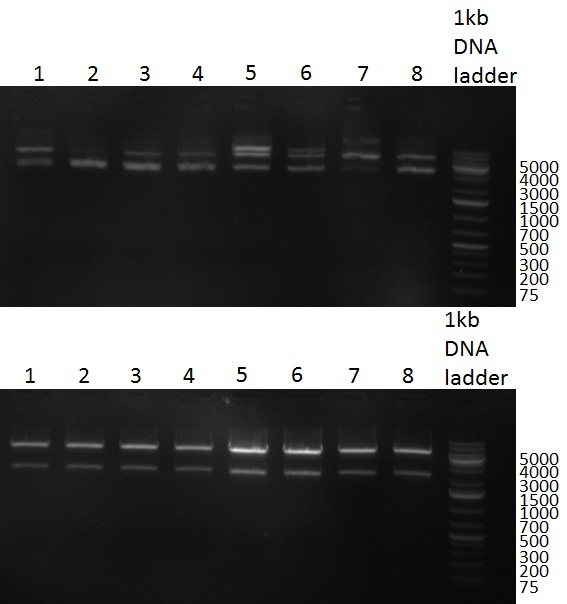
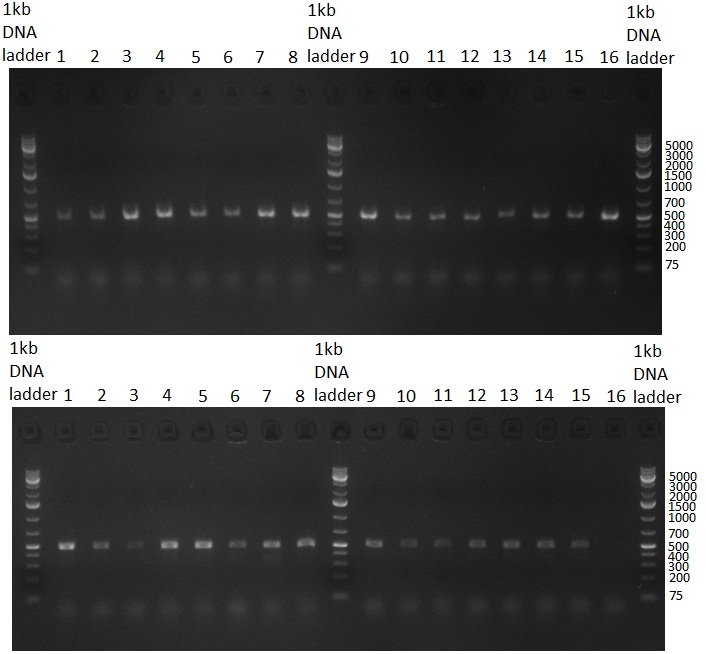
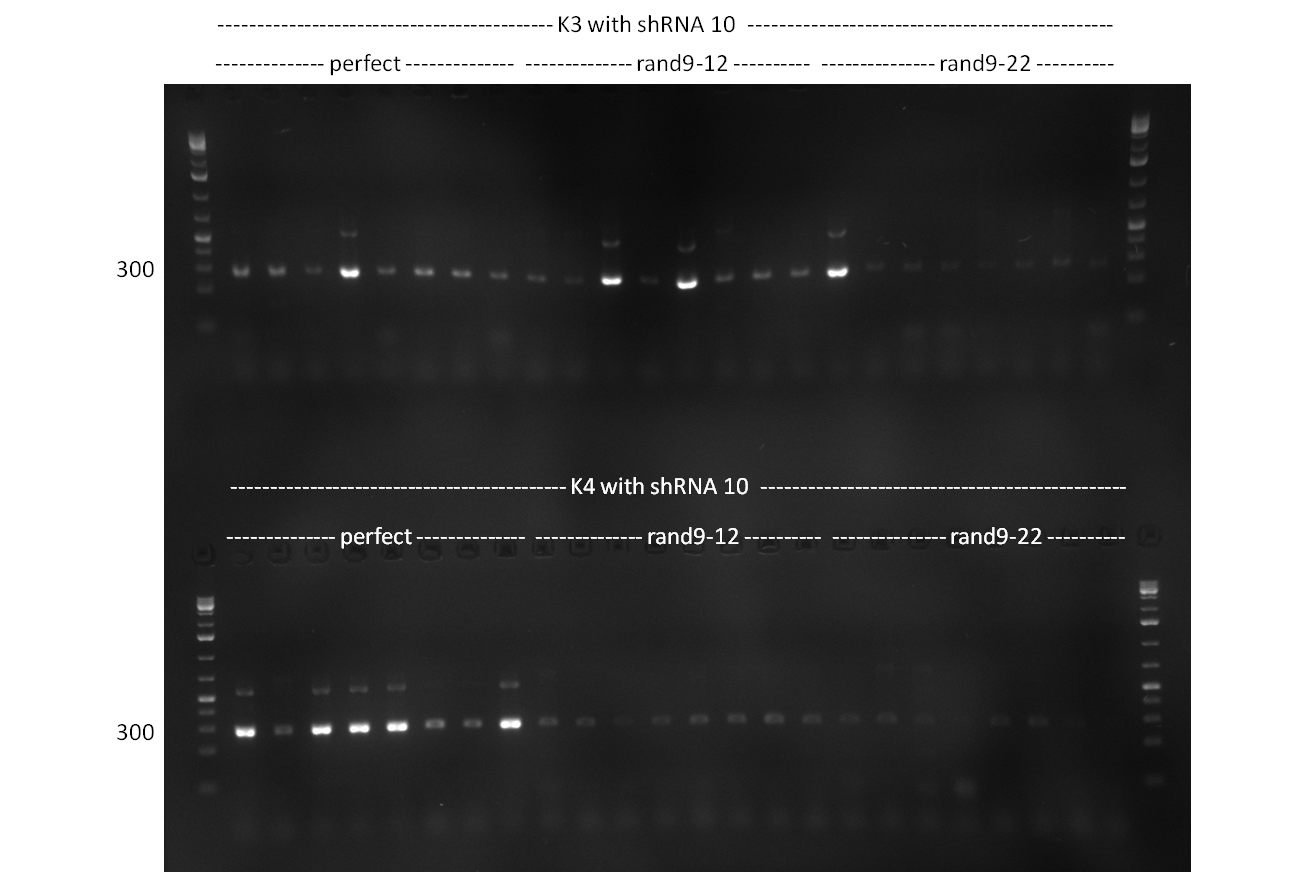
| + | [[Image:20101001_gel1_HD2010.png|thumb|500 px|right|Screening 1: shRNA10 single binding sites]] | ||
| + | [[Image:20101001_gel2_HD2010.png|thumb|500 px|right|Screening 2: shRNA10 single binding sites]] | ||
| + | |||
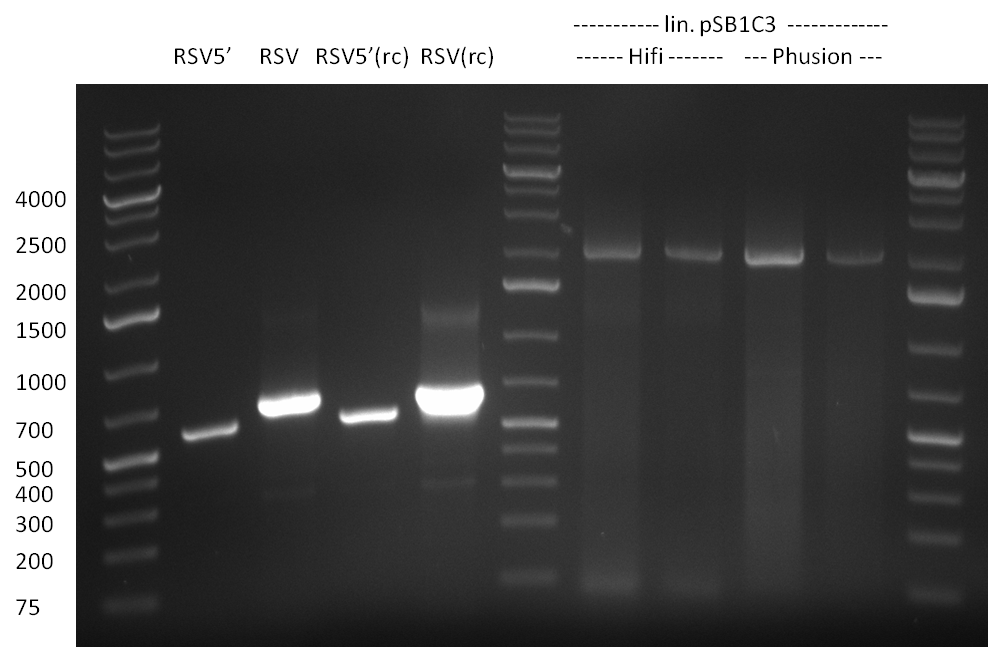
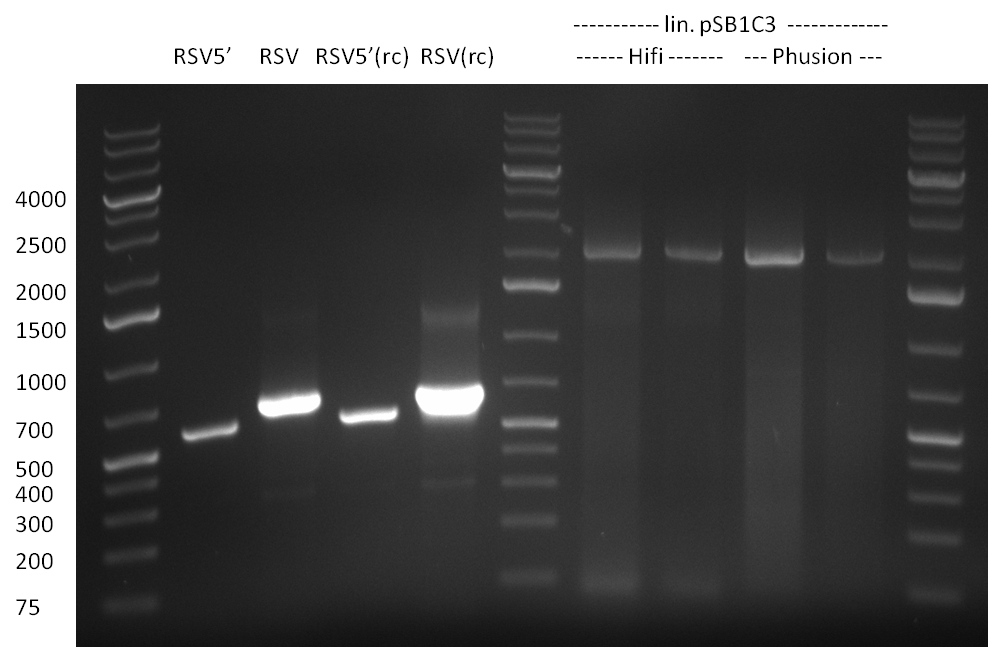
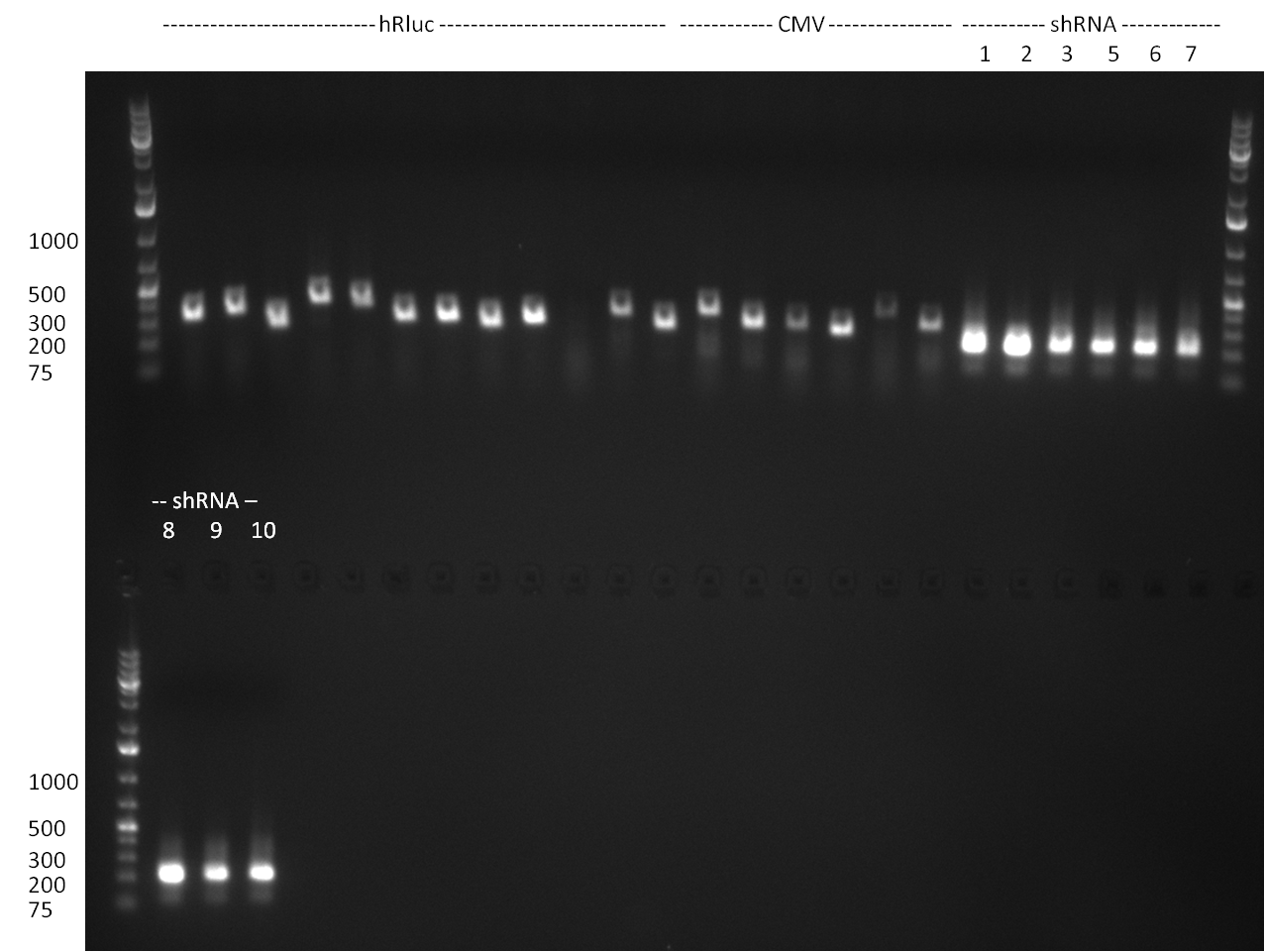
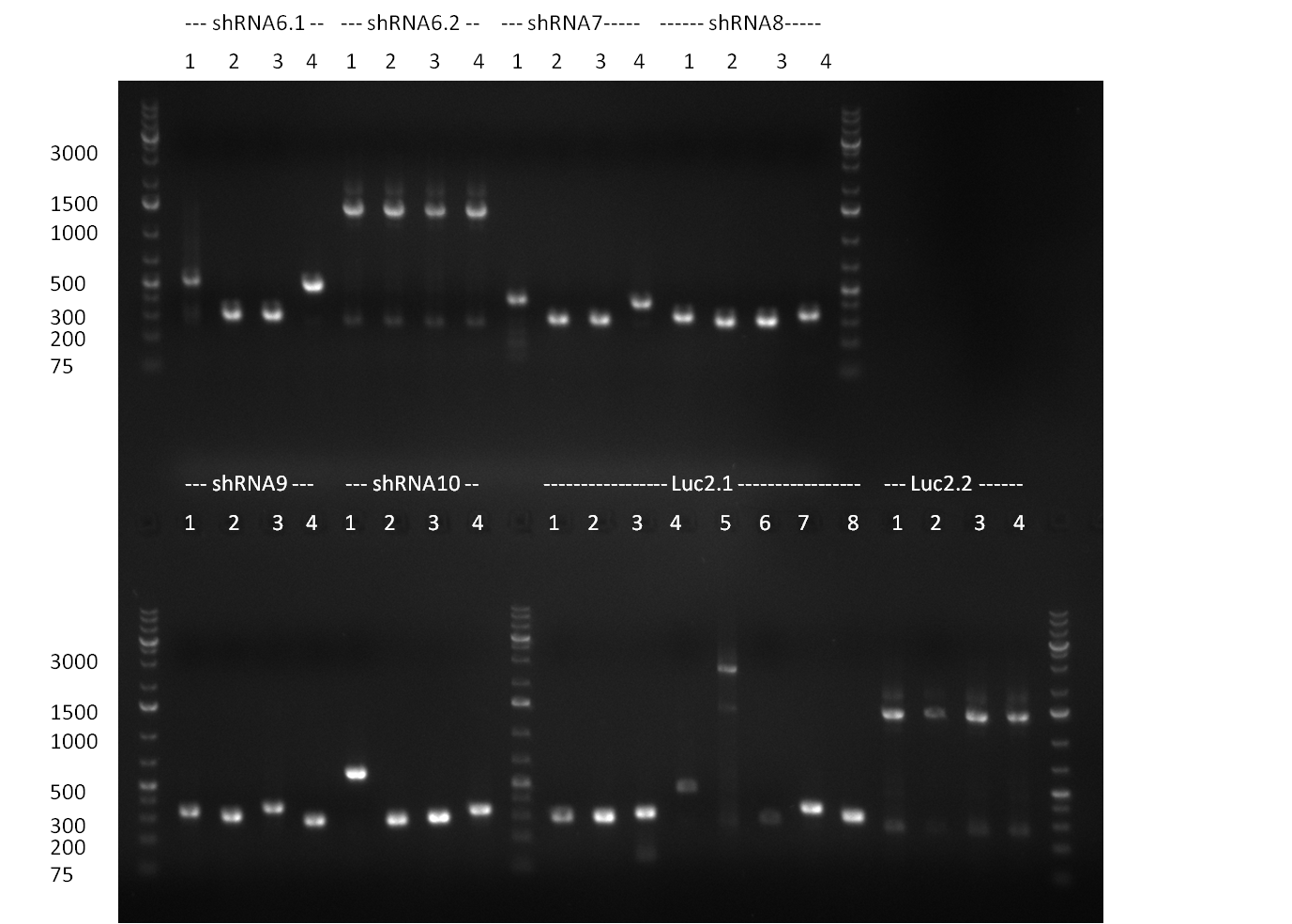
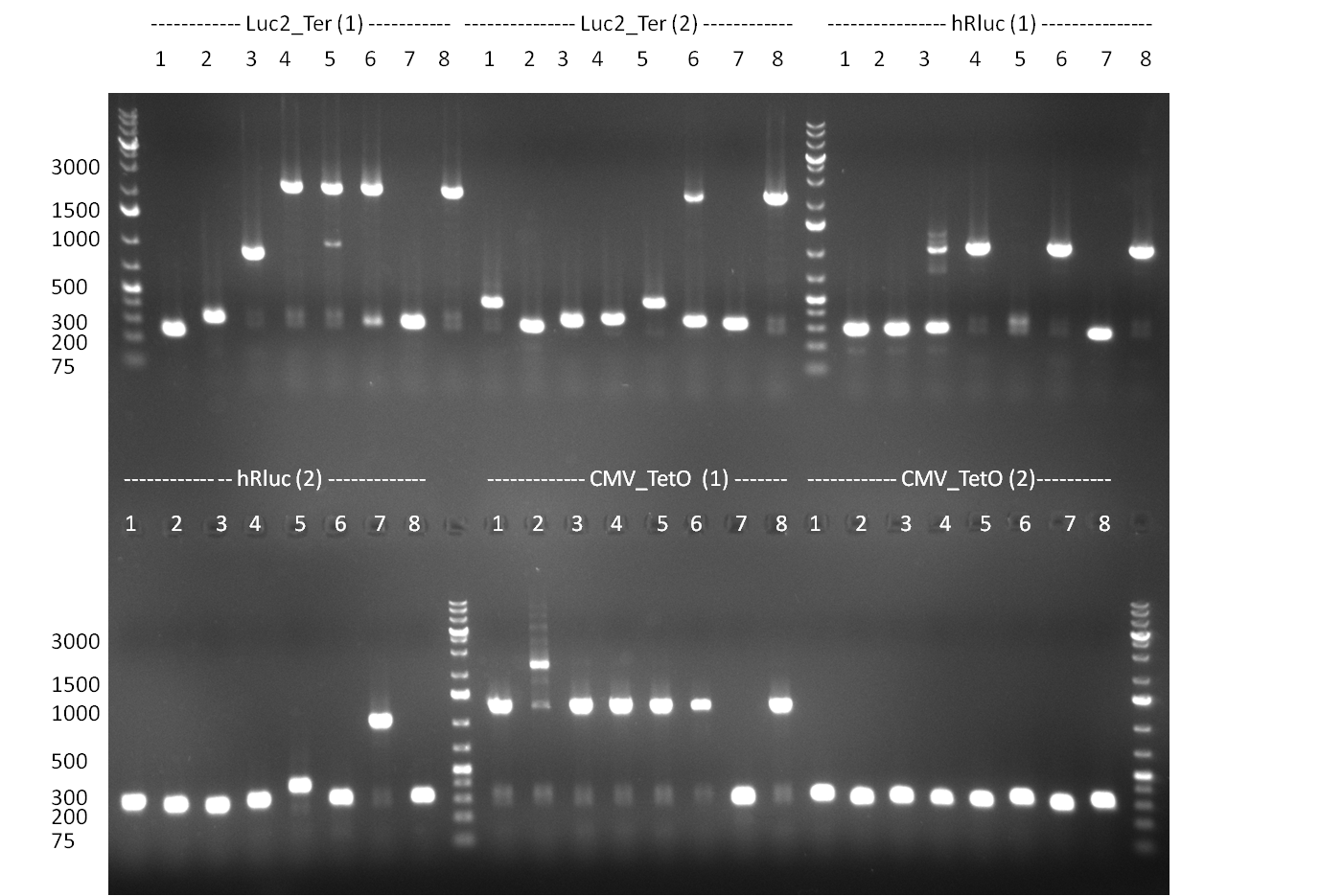
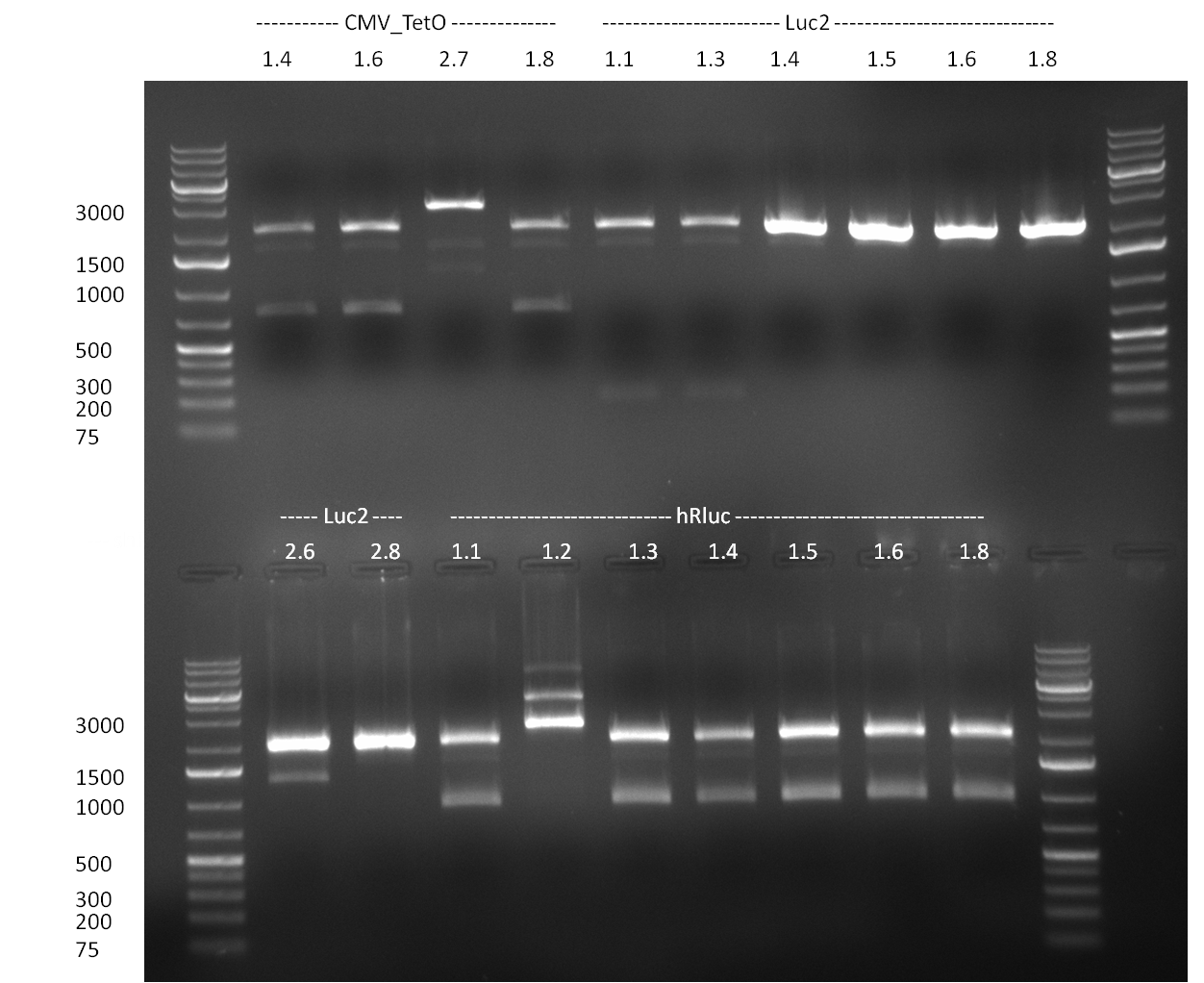
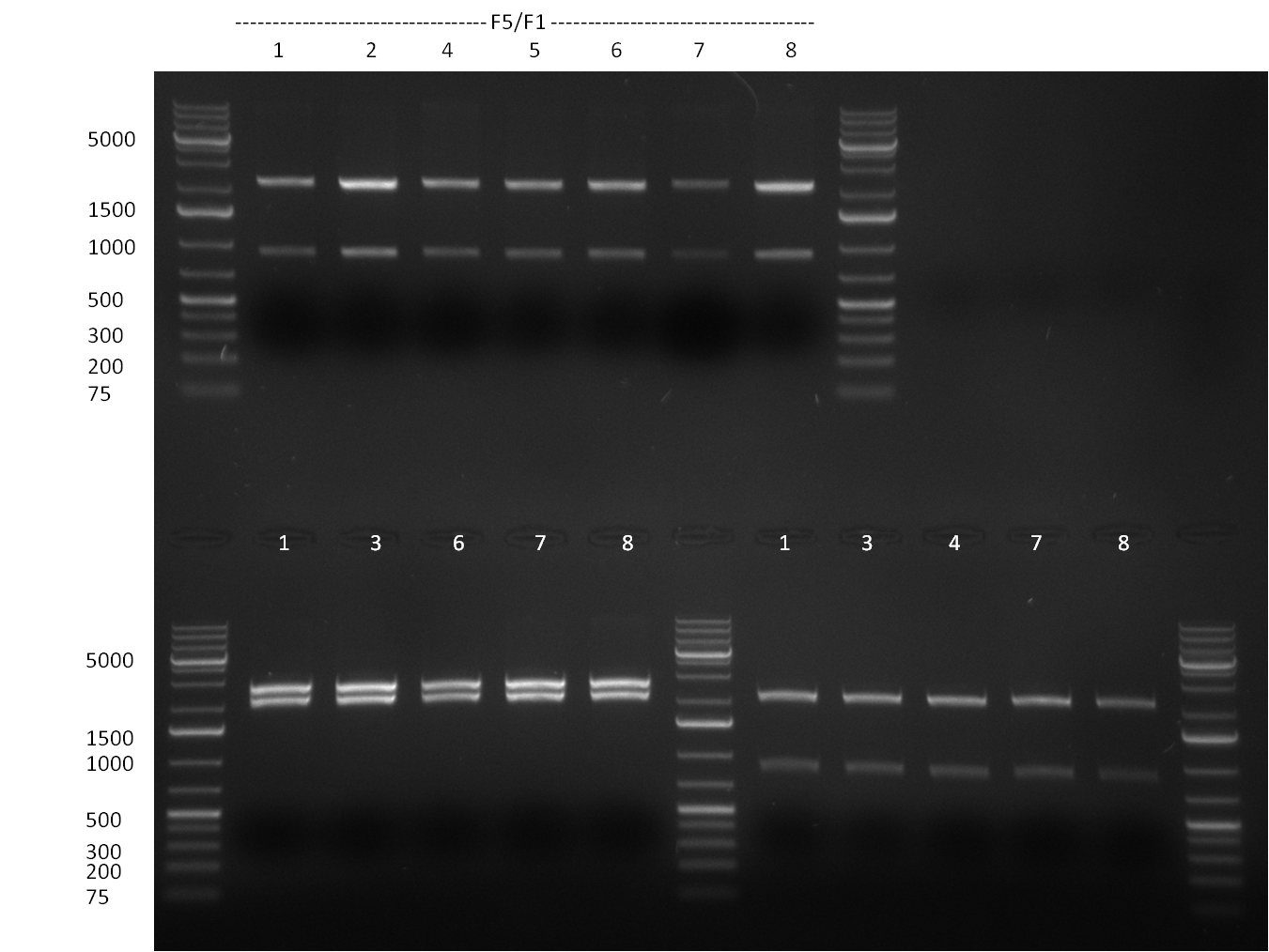
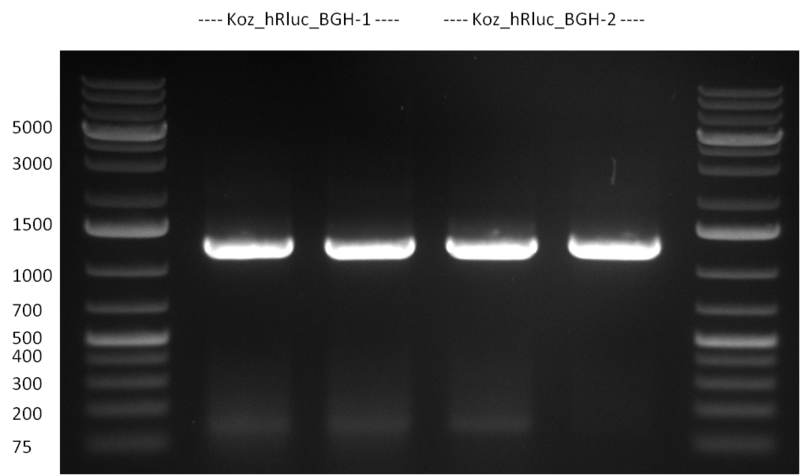
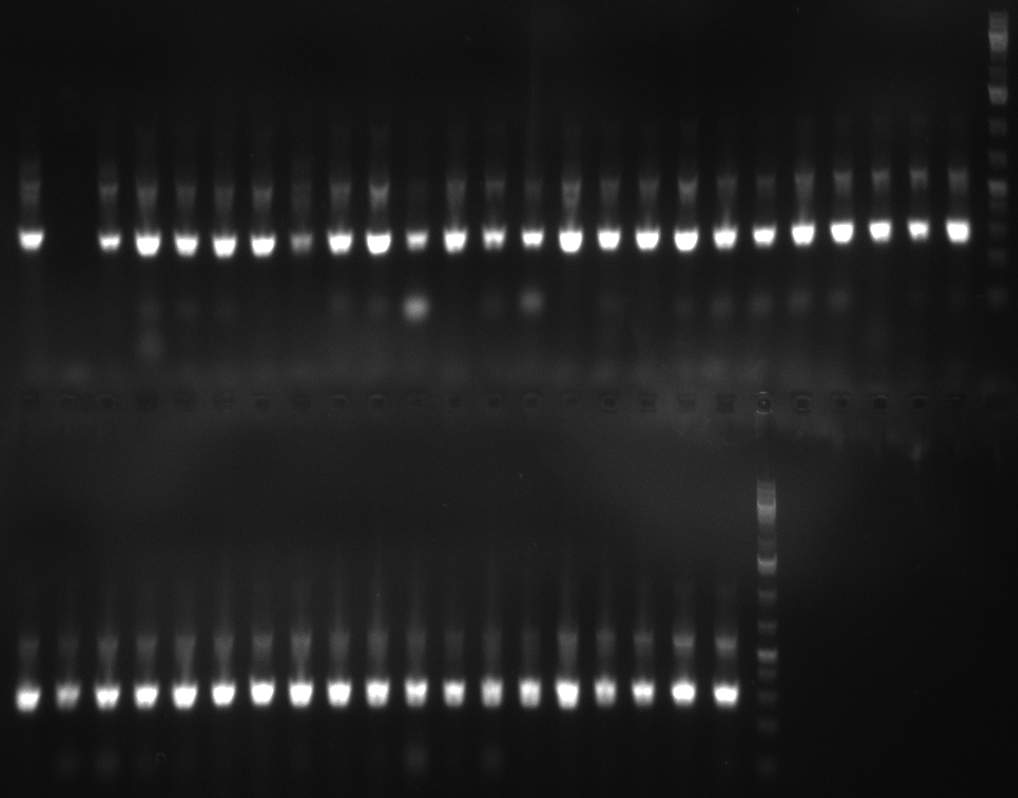
| + | *Screening of the binding sites cloned into the constructs K3 and K4 containing shRNA10 in pSB1A3 on the previous day. 16 colonies were picked for each perfect, rand9-12 and rand9-22 binding site and colony-PCR was performed according to the colony-PCR standard protocol (Primers Luc2_bs_screen_seq and sh10_ape_xma_screen_rc). The results are shown on the gels Screening1 and Screening 2. Indicated are the construct numbers (either K3 or K4, which differ in the promoter driving the Luc2 gene) and the binding site property (perfect, 9-12 randomized or 9-22 randomized). Almost all clones show a positive band, although only some have a really bright band at the size of ~ 250 bp. LB cultures for 10 clones for each constructs K3 and K4 were inocculated. | ||
| + | |||
| + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| + | |||
| + | = 02/10/2010 = | ||
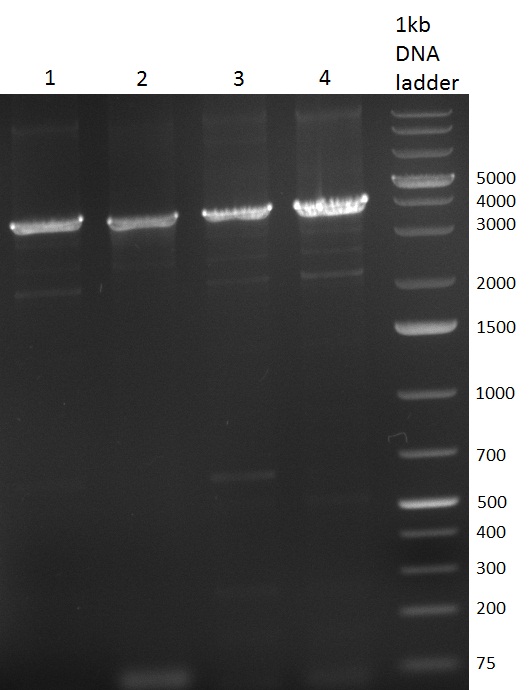
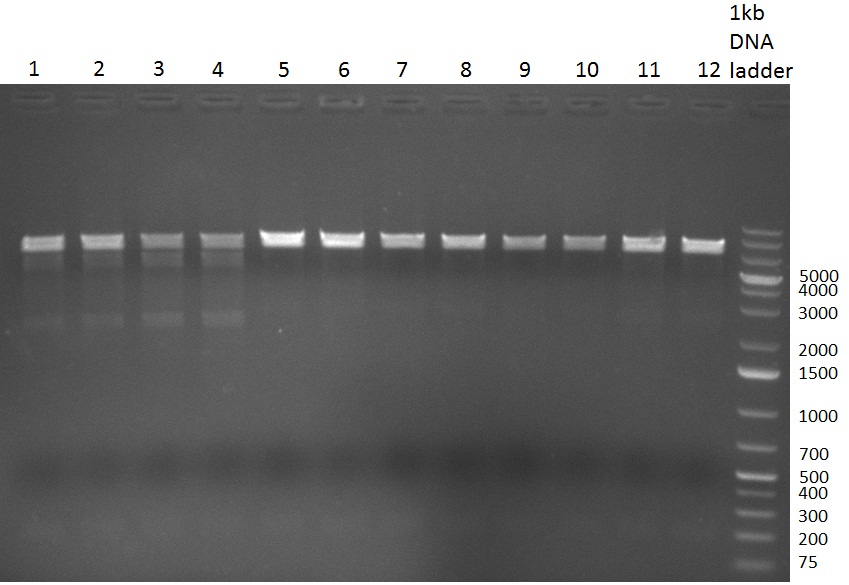
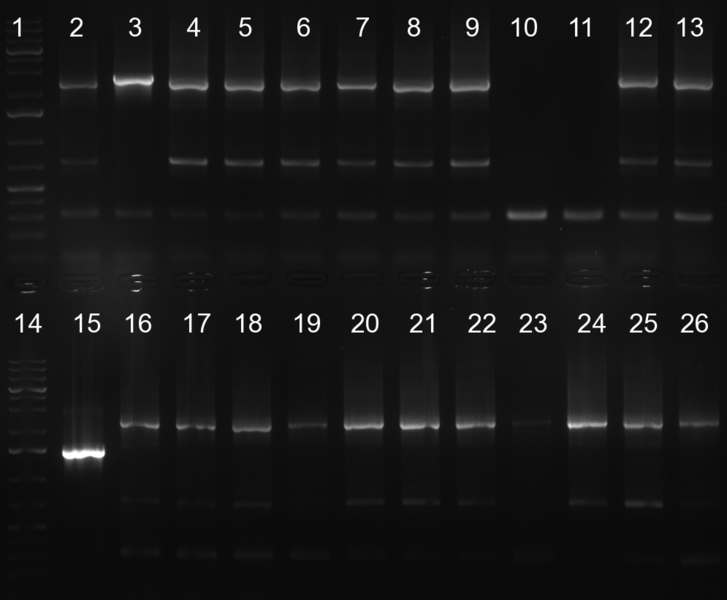
| + | [[Image:20101002 T1 T2 colonypcr.png|thumb|300px|right|Gel 101002-1. Colony PCR. All lanes with right amplified fragment except on lane 3, 10, 11, 15 and 23. Lane 1 & 14: 1kb Plus Ladder (Invitrogen)]] | ||
| + | |||
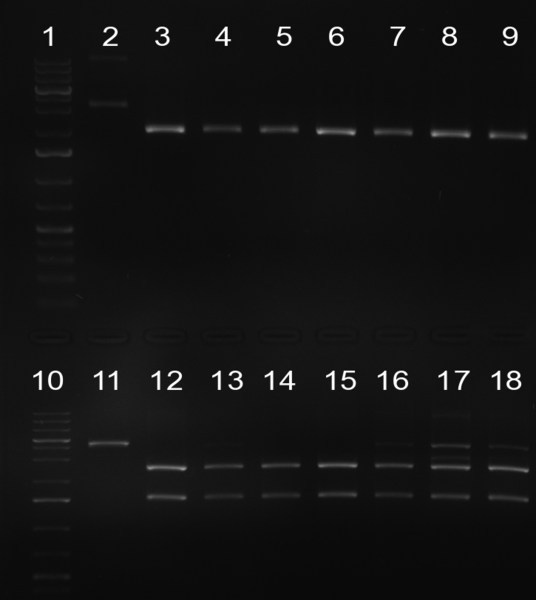
| + | [[Image:20101002 T1 T2 testdigest.png|thumb|300px|right|Gel 101002-2. Test digestions of repressor construct. Upper pattern: EcoRI & PstI. Lower pattern: BamHI. All bands revealing positive results except in the first column (negative control from lane 3 of gel 101002-1). Lane 1 and Lane 10: 1kb Plus Ladder (Invitrogen). Lane 16, 17, 18: incomplete digestion ]] | ||
| + | |||
| + | * Miniprep of the LB cultures (constructs K3 and K4 with shRNA10 and the corresponding binding sites) were done and used for transfection of Hek-293 cells | ||
| + | |||
| + | |||
| + | repressor construct:<br/> | ||
| + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of [[Igem2010/Main/synthetic miR Kit/October#02/10/2010 | previous day transformations]] | ||
| + | ** analysis, see gel 101002-1 | ||
| + | * twice test digestion of promising mini-preps, following [https://2010.igem.org/3A_Assembly standard protocol recommendations] <br/> | ||
| + | ** first digestion with EcoRI and PstI to check for insert size | ||
| + | ** second digestion with BamHI to check the insert | ||
| + | *** see gel 101002-2: some positive results will be send for sequencing and typed [[Igem2010/Main/Library/Constructs/pSMB miTrigger | T1 and T2]], respectively | ||
| + | **** moreover: usage of one construct for transfection into HEK cells | ||
| + | |||
| + | <br/><br/> | ||
| + | |||
| + | = 03/10/2010 = | ||
| + | repressor construct: <br/> | ||
| + | * sequencing results were fine | ||
| + | * digestion and purification of [[Igem2010/Main/Library/Constructs/pSMB miTrigger | T1 and T2]] with XmaI and XhoI to clone in binding sites (against ON-targets) following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ||
| + | * annealing of binding sites using oligos | ||
| + | ** D108 & D109: once binding site for miR122 | ||
| + | ** D110 & D111: twice binding site for miR122 right behind each other | ||
| + | *** protocol: | ||
| + | **** 5 µl of each oligo, 5 µl of Buffer 2 (NEB), 35 µl H<sub>2</sub>O | ||
| + | **** 95°C, 5' | ||
| + | **** cool down to room temperature for 2h | ||
| + | **** incubation on ice, 10' | ||
| + | **** use 1 µl for ligation with <nowiki>~</nowiki> 40 ng of backbone (i. e. digested [[Igem2010/Main/Library/Constructs/pSMB miTrigger | T1 and T2]]) | ||
| + | * ligation (including to negative controls with backbone only to check for re-ligation without SAP treatment) | ||
| + | * [https://2010.igem.org/Transformation transformation] and growth on selective plates (ampicillin) | ||
| + | |||
| + | = 04/10/2010 = | ||
| + | repressor construct:<br/> | ||
| + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of [[Igem2010/Main/synthetic miR Kit/October#03/10/2010 | previous day transformations]] | ||
| + | ** gel 101004-1 reveals almost positive samples | ||
| + | * submission of each a promising mini-prep for sequencing of binding site and correct insert | ||
| + | |||
| + | = 05/10/2010 = | ||
| + | repressor construct:<br/> | ||
| + | * <i>sequencing results</i> from [[Igem2010/Main/synthetic miR Kit/October#05/10/2010 | previous day]]: | ||
| + | ** once or twice the binding site in each of the T1 constructs, respectively | ||
| + | ** once the binding site for the T2 construct | ||
| + | *** re-trial of ligation for two binding sites in T2 | ||
| + | * planned transfection into HeLa and HUH cells (non-liver and liver to prove ON-targeting) in ratio 6:1 = repressor:operator(tuning) as recommended in the [http://tools.invitrogen.com/content/sfs/manuals/trexsystem_man.pdf T-Rex manual] | ||
| + | |||
| + | new approach: | ||
| + | * purification of PCR amplified shRNA-like miRNA against hAAT ("shhAAT") | ||
| + | * digestion of shhAAT and [[Igem2010/Main/Library/Constructs/pSMB miTrigger | Q2 and Q3]] with HindIII and AflII (for backbones sequential because of two inner AflII cutting sites) | ||
| + | * ligation in triplicates (using 70 ng of backbone and 10, 15 or 20 ng of insert, respectively) | ||
| + | * [https://2010.igem.org/Transformation transformation], over-night growth on selective agar plates (ampicillin) | ||
| + | |||
| + | = 06/10/2010 = | ||
| + | * waiting for colonies from [[Igem2010/Main/synthetic miR Kit/October#05/10/2010 | previous night]], which did not grow after all | ||
| + | |||
| + | = 07/10/2010 = | ||
| + | * repetition of shhAAT cloning | ||
| + | * touch-down PCR setup: 8.5 µl ddH<sub>2</sub>O, 0.5 µl template DNA (pcDNA5, 50 ng/µl), 0.5 µl each primer (i.e. [[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D7 & D16]], 10 µl Phusion PCR MasterMix (2x) | ||
| + | * touch-down PCR protocol: [[Igem2010/Main/Protocols/Phusion_PCR#shRNA-PCR Stefan M. protocol| like this]] but with increase of annealing temperature from 68°C to 61°C (i. e. 0.2°C per cycle) | ||
| + | * go on like [[Igem2010/Main/synthetic miR Kit/October#05/10/2010 | 05/10/2010]] but growth on freshly prepared plates | ||
| + | |||
| + | = 08/10/2010 = | ||
| + | * mini-prep and submission for sequencing for the only promising colony of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | Q2]] containing shhAAT for sequencing | ||
| + | |||
| + | = 09/10/2010 = | ||
| + | * sequencing [[Igem2010/Main/synthetic miR Kit/October#08/10/2010 | yesterday]] was fine but construct contains TetO<sub>2</sub>, thus: maybe Q2 and Q3 were confused | ||
| + | ** that would explain why measurements with TetR constructs are not working properly even though sequences were fine | ||
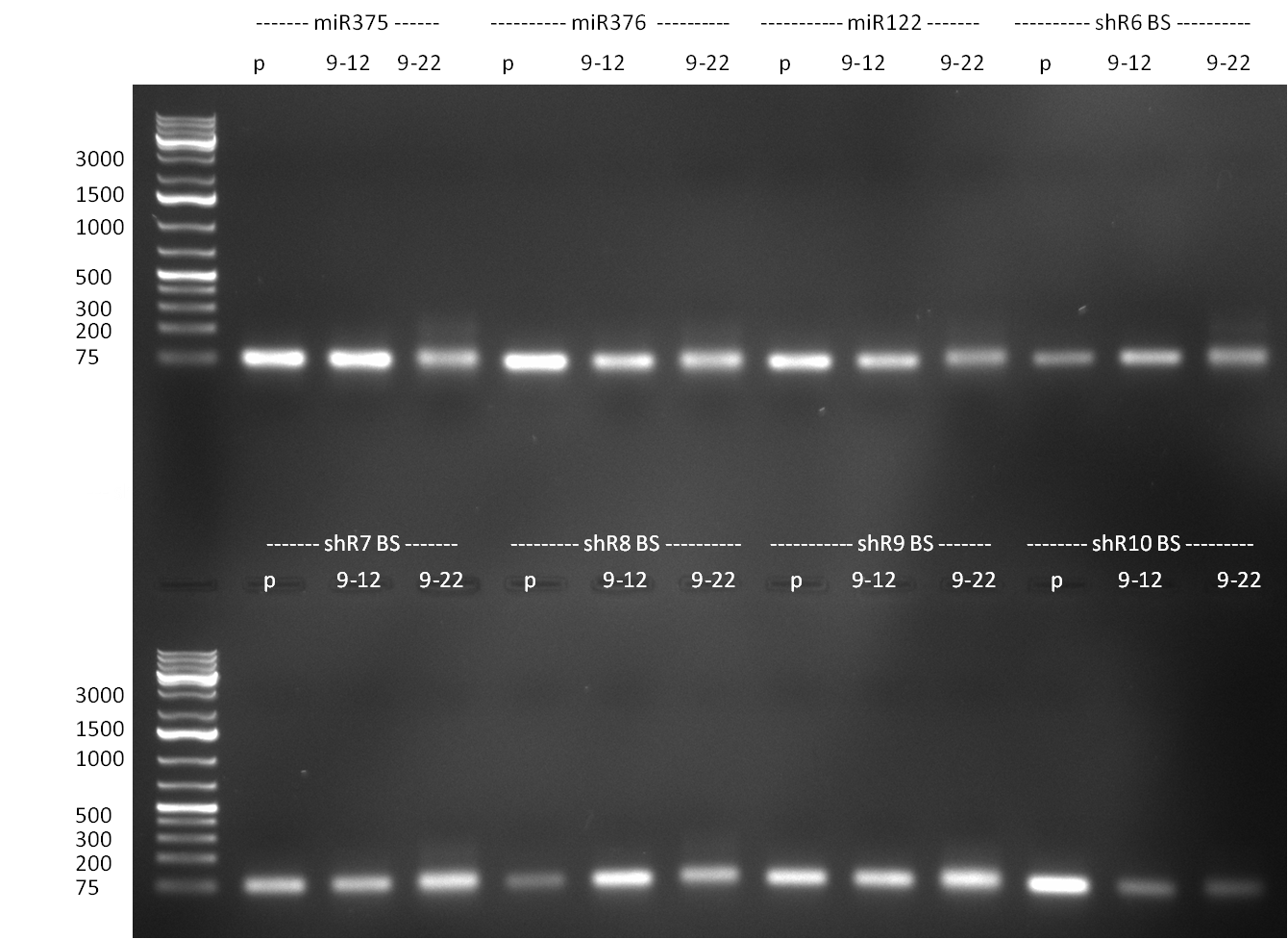
| + | * cloning of binding sites for shhAAT (perfect binding site generated via annealing, and randomized ones PCR amplified) | ||
| + | ** touch-down PCR setup: 17 µl ddH<sub>2</sub>O, 4 µl each oligo (i.e. respective forward primer + second strand oligo[[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D121]]), 25 µl Phusion PCR MasterMix (2x) | ||
| + | *** touch-down PCR protocol: [[Igem2010/Main/Protocols/Phusion_PCR#shRNA-PCR Stefan M. protocol| like this]] but with increase of annealing temperature from 70°C to 62°C (i. e. 0.5°C per cycle, 16x) and four additional cycles with 62°C | ||
| + | ** annealing of perfect binding against shhAAT sites with an oligo-based strategy following the protocol from the [[Igem2010/Main/synthetic miR Kit/October#03/10/2010 | 3<sup>rd</sup> October]] | ||
| + | ** digestion of PCR amplified binding sites with AgeI and XhoI, digestion of Q2 backbone containing the shhAAT with XhoI and XmaI, afterwards heat inactivation, nucleotide removal and ligation following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ||
| + | ** [https://2010.igem.org/Transformation transformation] and over-night growth on selective agar plates (ampicillin) | ||
| + | |||
| + | = 10/10/2010 = | ||
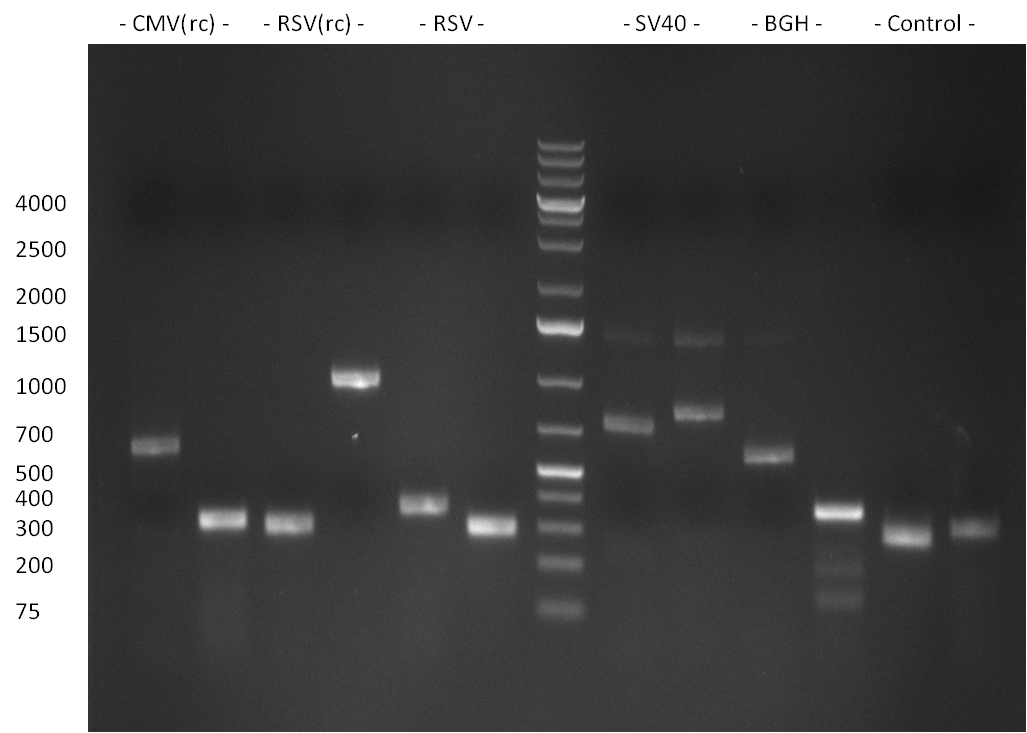
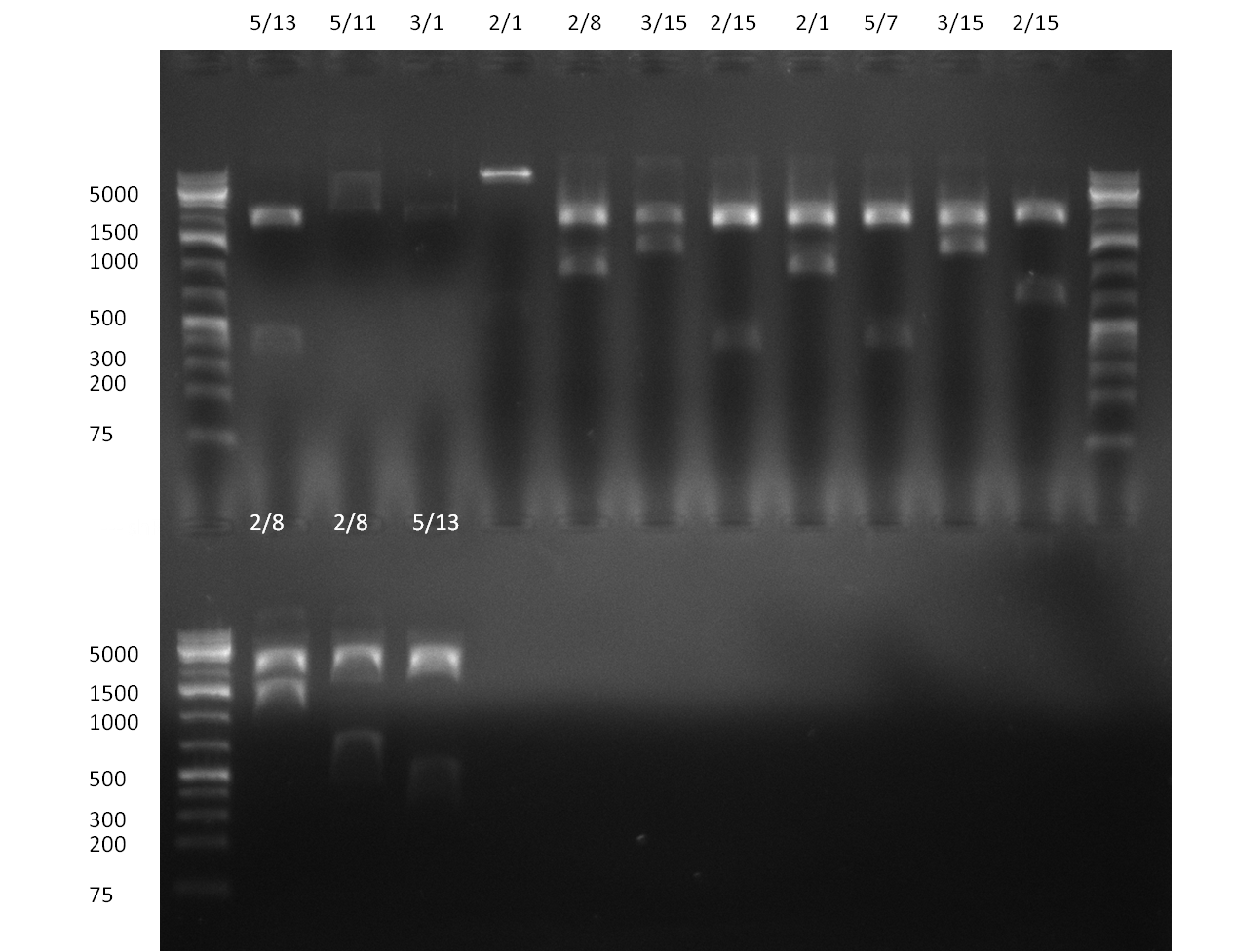
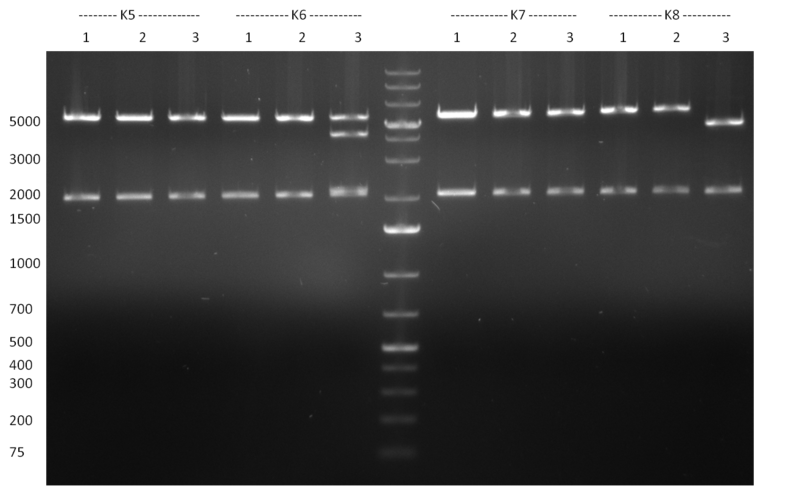
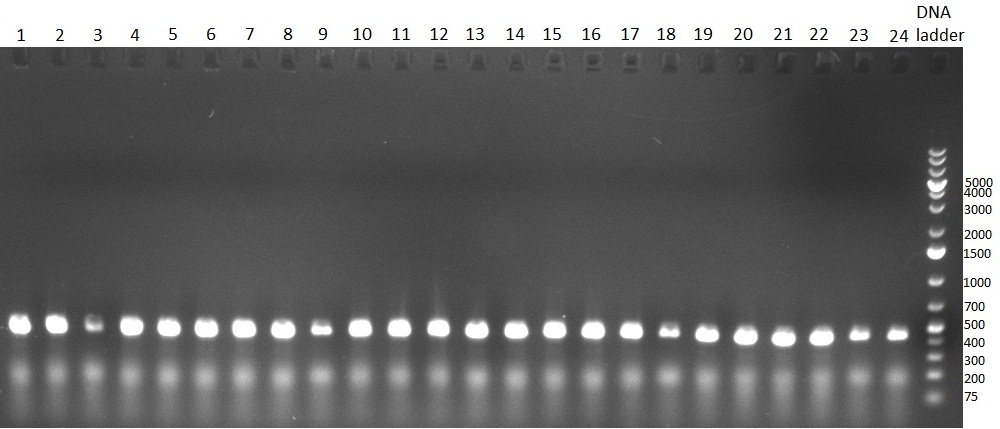
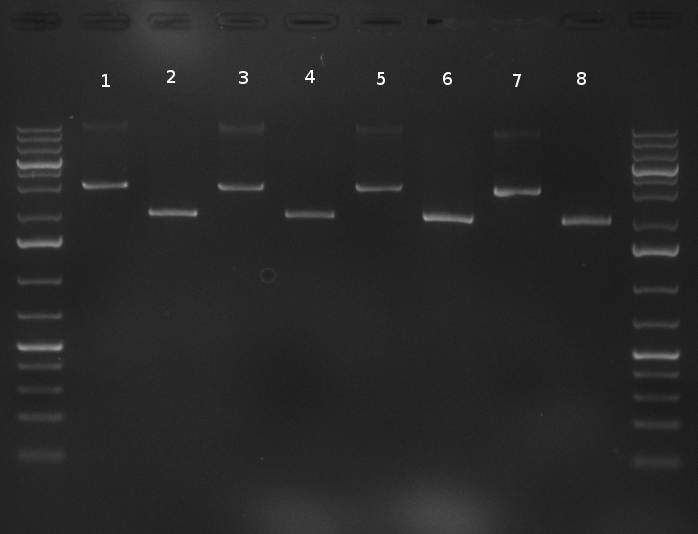
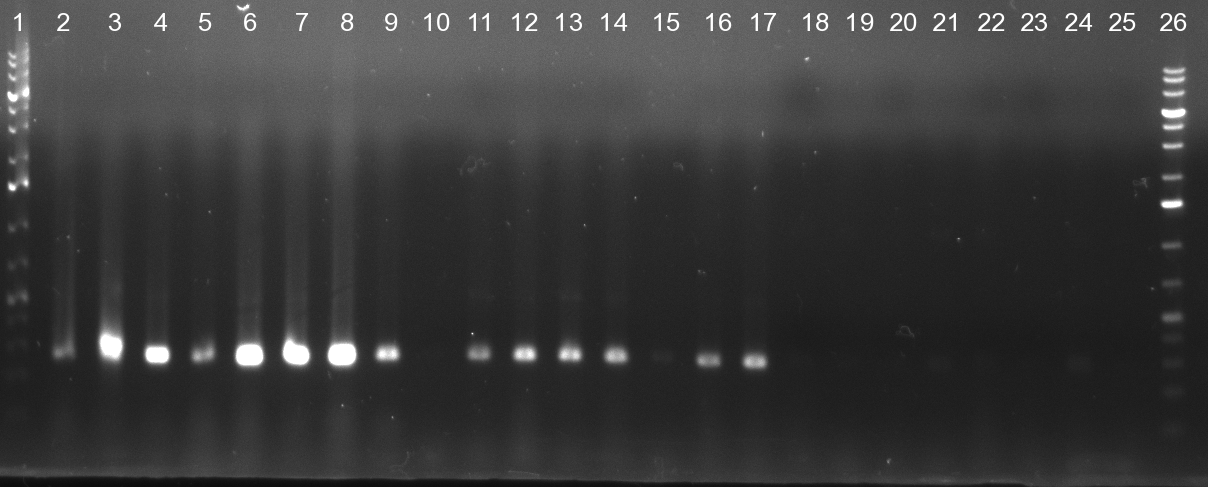
| + | [[Image:20101010 colonypcr shhAAT p 12 22 bs LOLA.png|thumb|300px|right|Gel 101010-1. Colony PCR for construct containing shhAAT and binding sites. Expected is an amplified fragment a bit smaller than 300 bp which can be seen on lane 3, 4, 6, 7, 8 (perfect binding sites), 12, 13, 14, 16 and 17 (randomized binding sites). Lane 1 and Lane 26: 1kb Plus Ladder (Invitrogen)]] | ||
| + | * colony pcr reveals some positive clones (see gel 101010-1) from [[Igem2010/Main/synthetic miR Kit/October#09/10/2010 | previous day]] | ||
| + | ** inoculation of 5 ml LB Amp cultures for mini-prep over night | ||
| + | |||
| + | = 11/10/2010 = | ||
| + | * mini-prep of ten cultures from [[Igem2010/Main/synthetic miR Kit/October#10/10/2010 | yesterday]] and direct transfection to test the system | ||
| + | ** amount: each 100 µl (c=2.5 ng/µl) | ||
| + | <br/> | ||
| + | |||
| + | measurement construct for single binding sites:<br/> | ||
| + | * PCR-based generation of binding sites against miR122 (perfect and randomized) | ||
| + | ** touch-down PCR setup: | ||
| + | *** 17 µl ddH<sub>2</sub>O, | ||
| + | *** 4 µl each oligo (i.e. respective forward primer [[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D71, D62 or D5]] + second strand oligo[[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D13]]), | ||
| + | *** 25 µl Phusion PCR MasterMix (2x) | ||
| + | ** touch-down PCR protocol: | ||
| + | *** [[Igem2010/Main/Protocols/Phusion_PCR#shRNA-PCR Stefan M. protocol| like this]] but with increase of annealing temperature from 70°C to 62°C (i. e. 0.5°C per cycle, 16x) | ||
| + | *** and four additional cycles with 62°C, elongation time only 10 seconds | ||
| + | * digestion of generated binding sites and the pSMB_miMeasure backbone with EcoRI and PstI, then nucleotide removal | ||
| + | * ligation with <nowiki>~</nowiki> 70 ng vector and between 7.5 ng and 15 ng insert | ||
| + | * [https://2010.igem.org/Transformation transformation] | ||
| + | |||
| + | =12/10/2010= | ||
| + | measurement construct for single binding sites: | ||
| + | * colony pcr of each eight of load of colonies + two negative controls (sum: 50 samples) | ||
| + | ** protocol: 16 cycles touch-down pcr from 70°C to 62°C of annealing temperature, then 19 additional cycles with 62°C annealing temperature | ||
| + | *** elongation time: 90<nowiki>''</nowiki> | ||
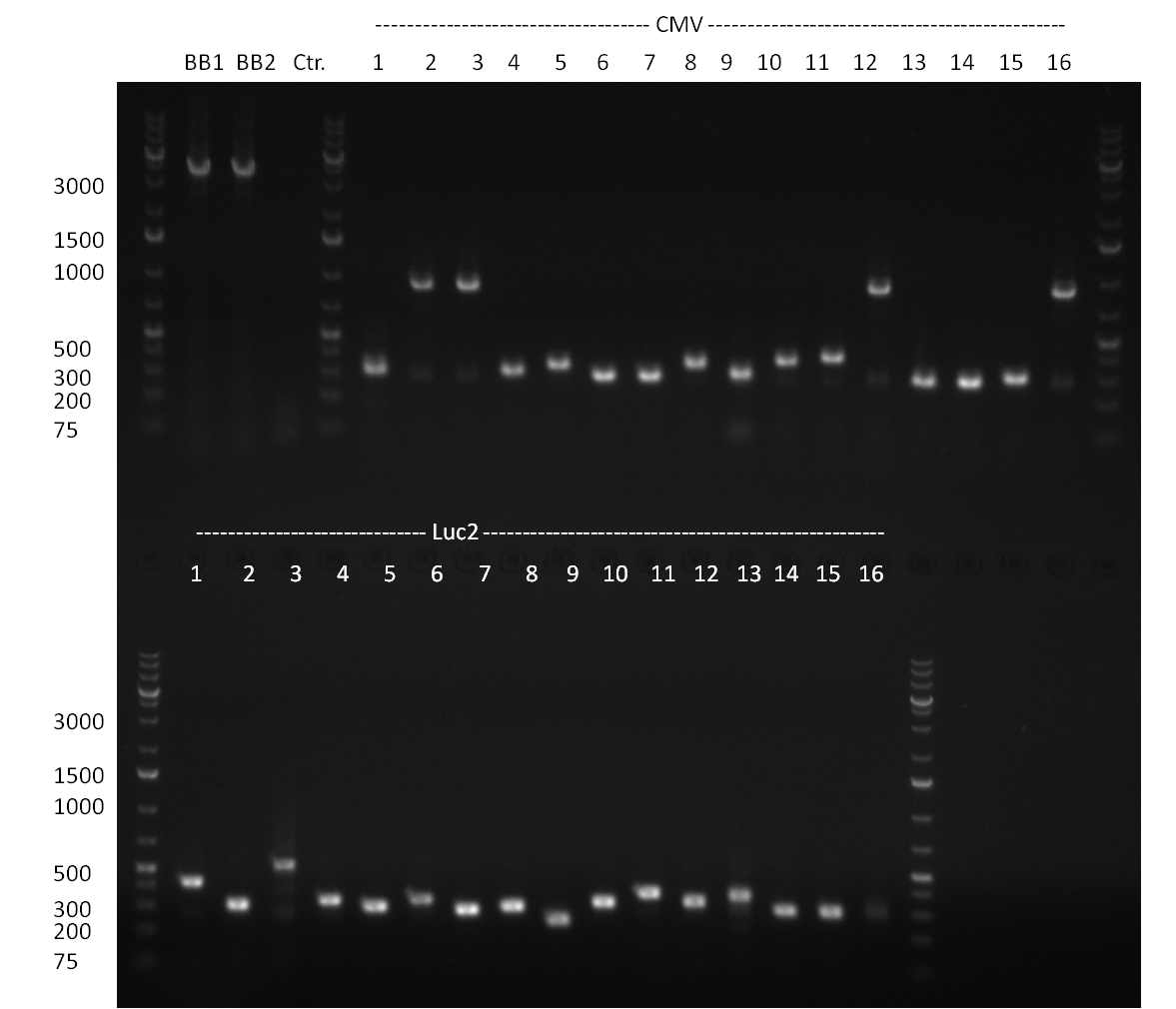
| + | ** <u>no positive</u> results, maybe due to insufficient screening primers (one for BBb standard, the other one for CMV resulting in an 1300 bp fragment) | ||
| + | *** as a consequence: repetition of entire cloning with [http://www.fermentas.com/templates/files/tiny_mce/coa_pdf/coa_ef0511.pdf SAP] treated backbone [[Igem2010/Main/synthetic miR Kit/October#12/10/2010| tomorrow]] | ||
| + | |||
| + | =13/10/2010= | ||
| + | measurement construct for single binding sites: <br/> | ||
| + | * repetition of cloning from [[Igem2010/Main/synthetic miR Kit/October#11/10/2010 | 11/10/2010]] | ||
| + | * <u>note:</u> vector concentration of pSMB_miMeasure is okay but after digestion and nucleotide removal it is always pretty low | ||
| + | ** (loss of 75% of DNA on average whereas other DNA yields to 90% of purified DNA as compared to digested amount) | ||
| + | |||
| + | =14/10/2010= | ||
| + | * cloning of 47 - 58 into pSB_H1 | ||
| + | * cloning of tuning constructs for in vivo measurement: | ||
| + | **cloning of binding sites of miRNA haat with perfect and several imperfect binding | ||
| + | **oligo annealing following the standard protocol | ||
| + | ** digestion of backbone pbsU6 with NotI and XhoI | ||
| + | ** ligation and transformation | ||
| + | |||
| + | =15/10/2010= | ||
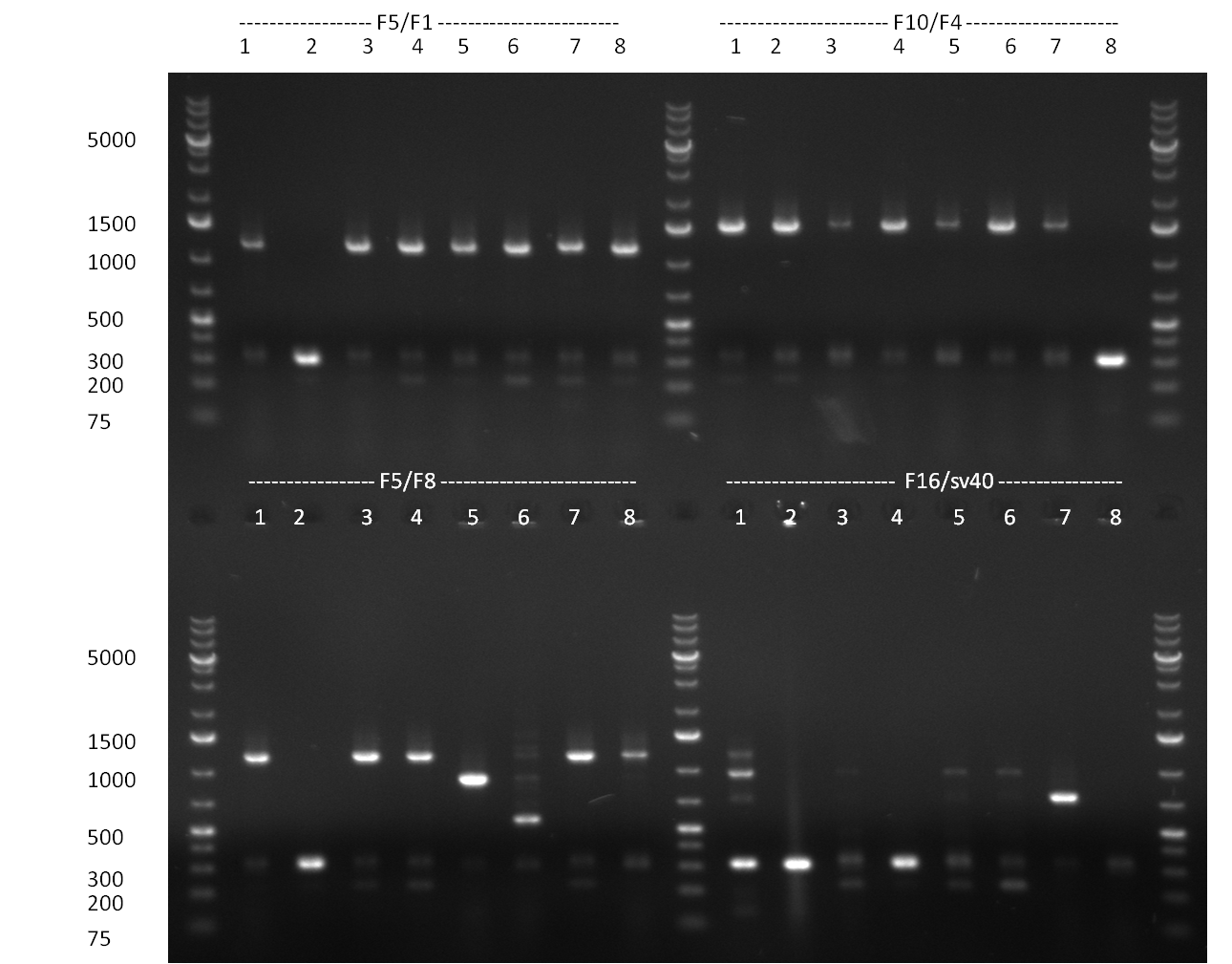
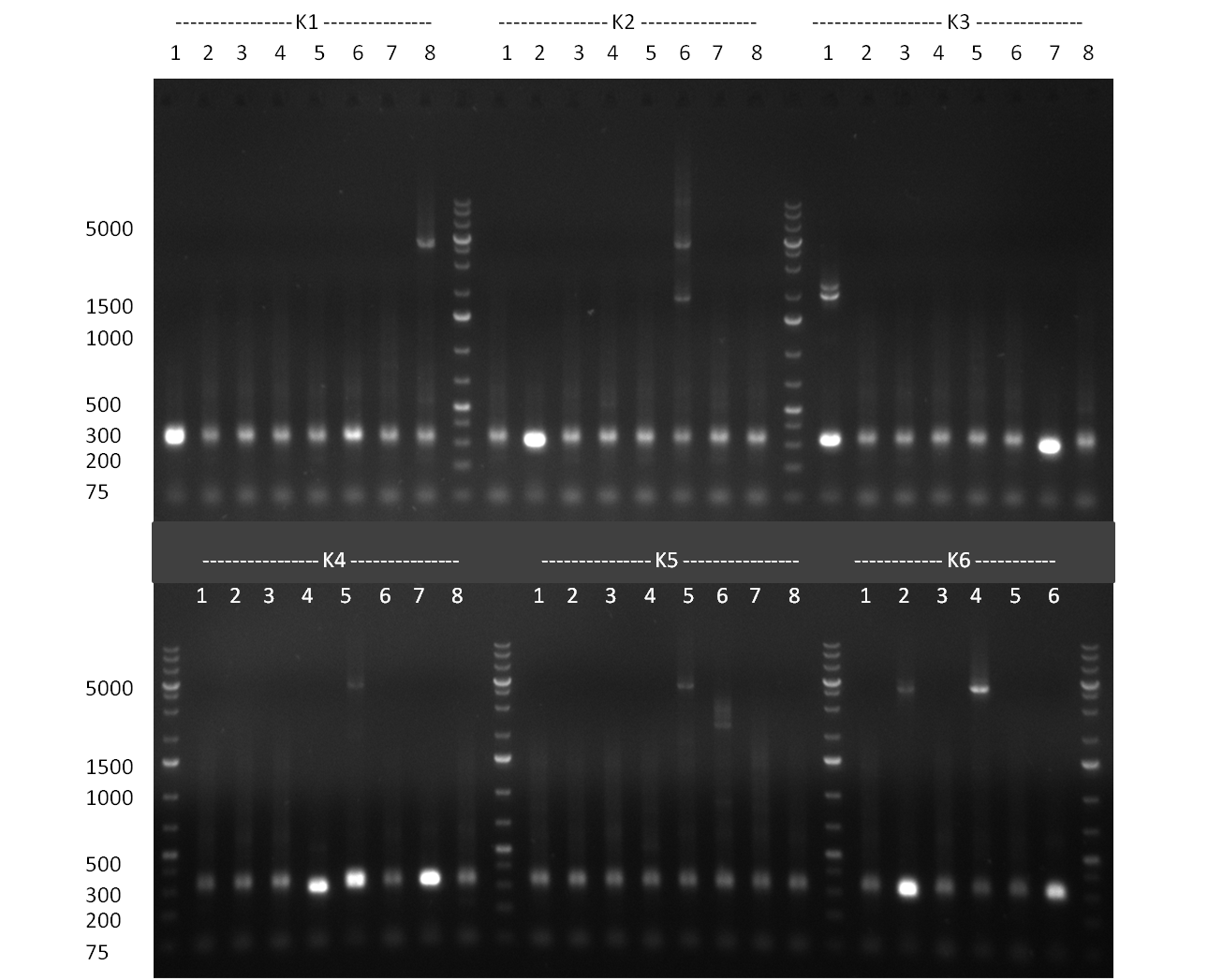
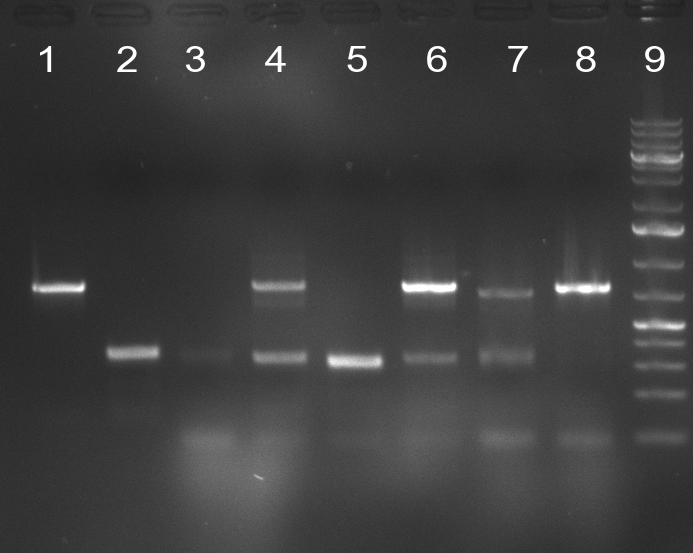
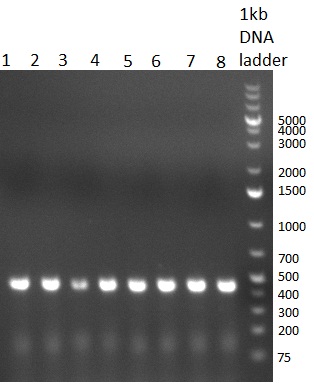
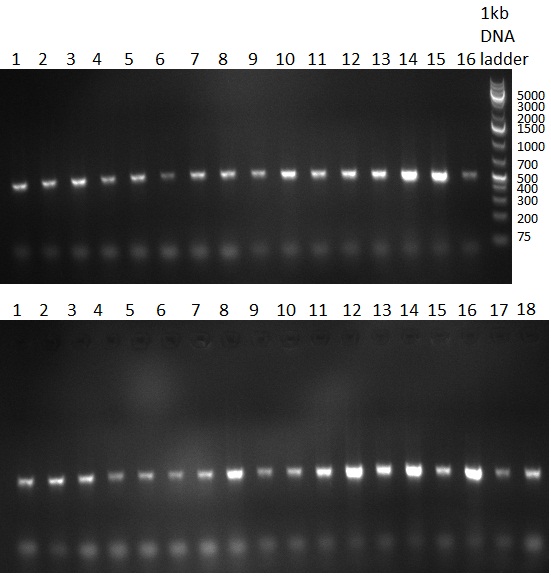
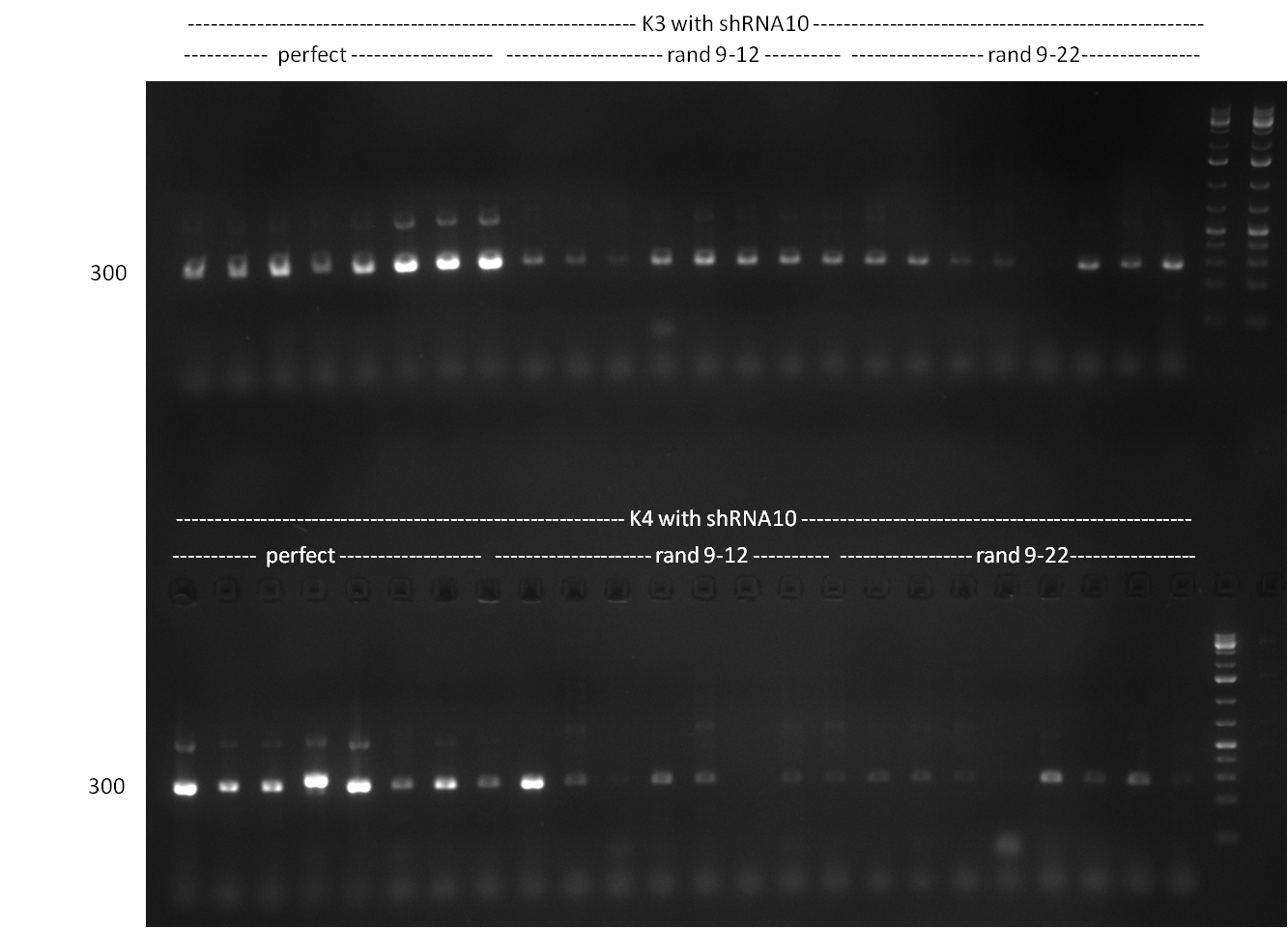
| + | [[Image:20101014_colonypcr_M1_M11anno.jpg|thumb|300px| Screening of M1 to M11 constructs, 4 clones were picked from each plate. 300bp band marks positive clones]] | ||
| + | [[Image:20101014_colonypcr_M12_M22anno.jpg|thumb|300px| Screening of M12 to M22 constructs, 4 clones were picked from each plate. 300bp band marks positive clones]] | ||
| + | [[Image:20101014_colonypcr_mimeasure122BS_p_12_22anno.jpg|thumb|300px| Screening of measurement standard with perfect, 9-12 and 9-22 randomized binding sites. constructs, 8 clones were picked from each plate. 300bp band marks positive clones]] | ||
| + | |||
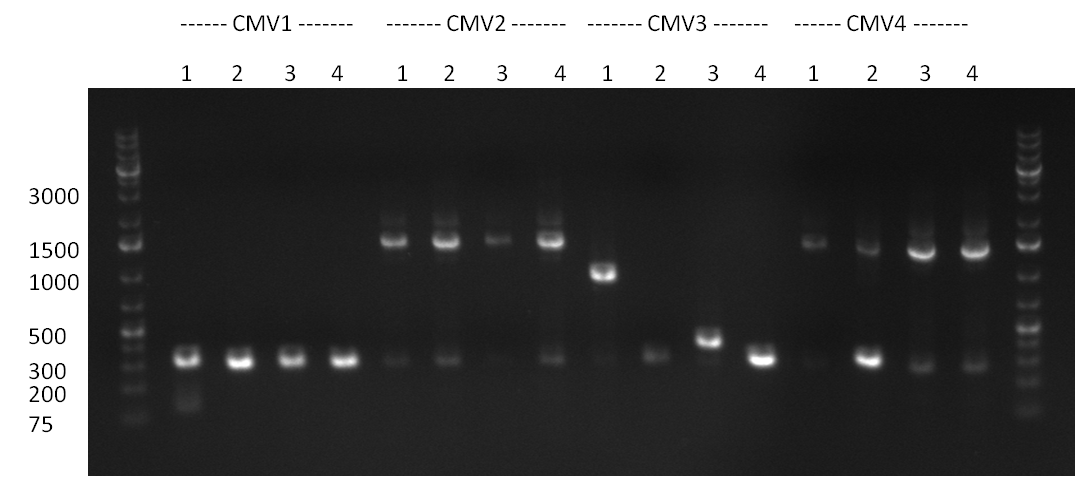
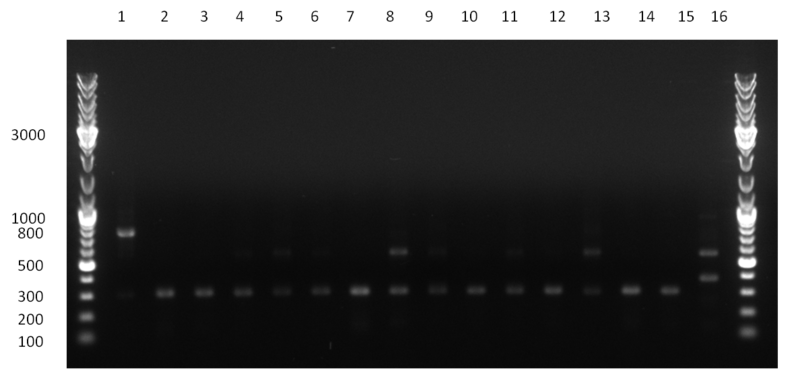
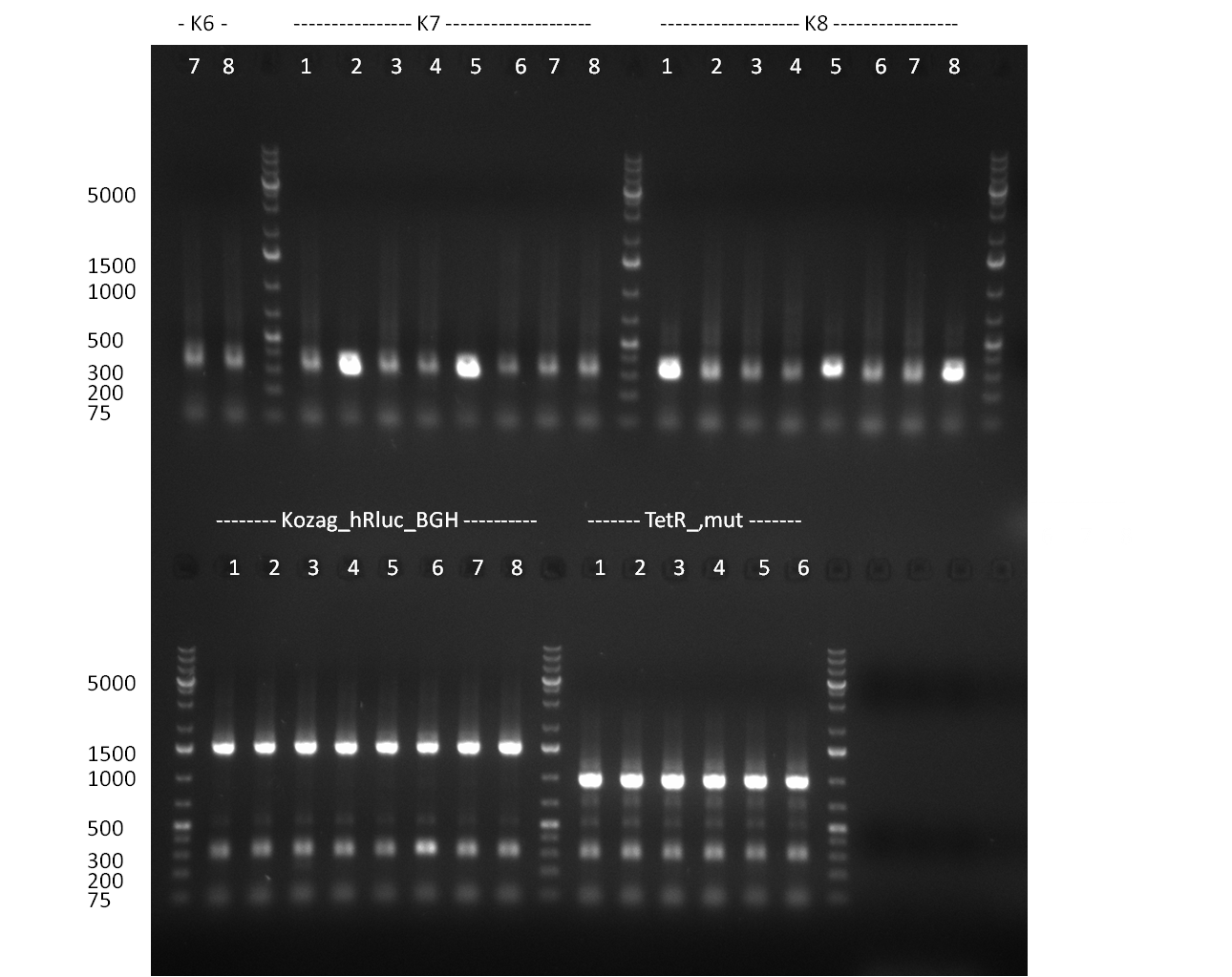
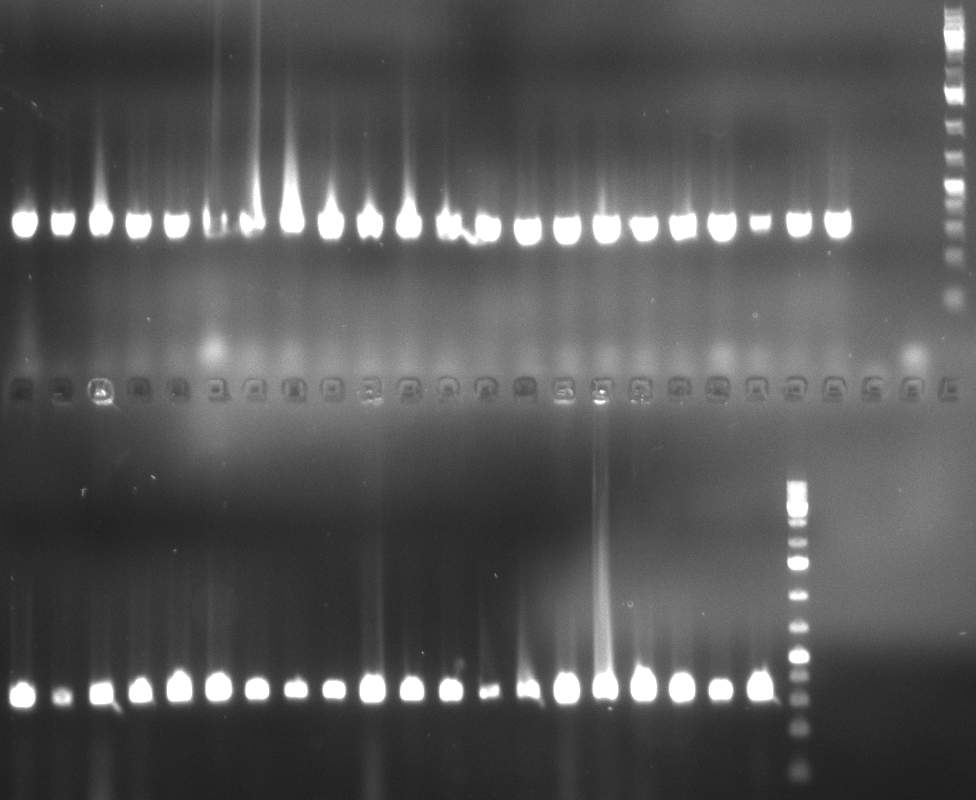
| + | * screening of plates M1 to M11 of the transformation with the construct pBSU6_SV40_Luc2 with 11 different binding sites for sh hAAT. Four clones from each plate were screened by colony pcr, positives give a band at 300bp. Primers Luc2_bs_screen_seq_fw and reverse primerse are the reverse binding site oligos for the binding sites. | ||
| + | ** inoculation of miniprep cultures for one positive from each plate. | ||
| + | |||
| + | * transformation of miRsAg and mir122 expression constructs | ||
| + | |||
| + | * screening of measurement standart cloning from 13th with perfect, 9-12 randomized and 9-22 randomized binding sites for miRNA 122. primers mir122_mimeasure_screen_rev and pSMB_miMeasure_screen_fw. Positives give a band at 300bp. | ||
| + | ** inoculation of miniprep cultures for positive clones | ||
| + | |||
| + | * screening of plates M12 to M22 of the transformation with the measurement plasmid with 11 different binding sites for miRsAg. Four clones from each plate were screened by colony pcr, positives give a band at 300bp. Primers pSMB_miMeasure_Sequencing_rev and forward primerse are the forward primer oligos for the binding sites. | ||
| + | ** inoculation of miniprep cultures for one positive from each plate. | ||
| + | |||
| + | =16/10/2010= | ||
| + | * mini Prep of mimeasure colonies (4 each) | ||
| + | * mini prep of haat constructs | ||
| + | * inoculation of miRsAg and mir122 expression constructs | ||
| + | * Maxi Preps of constructs M1 till M10 | ||
| + | * inoculation for Maxipreps of Adenovirus 5, AAV8 rep and cap genes and pBSU6 H1 haat construct | ||
| + | * inoculation for Maxiprep of constructs M11 an M23-29 | ||
| + | * cloning of on targeting constructs with a new strategy... | ||
| + | |||
| + | =17/10/2010= | ||
| + | * annealing of eleven sAg binding sites using oligos | ||
| + | *** protocol: | ||
| + | **** 5 µl of each oligo, 5 µl of Buffer 2 (NEB), 35 µl H<sub>2</sub>O | ||
| + | **** 95°C, 5' | ||
| + | **** cool down to room temperature for 2h | ||
| + | **** incubation on ice, 10' | ||
| + | **** use 1 µl for ligation with <nowiki>~</nowiki> 40 ng of backbone (i. e. with NotI and XhoI digested pBS_U6 containing hAAt cDNA) | ||
| + | * [https://2010.igem.org/Transformation transformation] and growth on selective plates (ampicillin) | ||
| + | |||
| + | = 18/10/2010 = | ||
| + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 4 colonies for each plate of [[Igem2010/Main/synthetic miR Kit/October#17/10/2010 | previous day transformations]] using reverse binding site oligo and forward hAAT primer expecting a fragment round about 1300 bp | ||
| + | ** gel 101018-1 reveals almost positive samples except two (data not yet shown) | ||
| + | |||
| + | = 19/10/2010 = | ||
| + | * mini-preps of cloned shAAT constructs | ||
| + | * dilution and co-transfection with miRsAg expressing construct into HeLa cells for ELISA measurements | ||
{{:Team:Heidelberg/Pagemiddle}} | {{:Team:Heidelberg/Pagemiddle}} | ||
| + | '''[[Igem2010/Main|⇐ Main Page]] | ||
| + | |||
| + | |||
| + | {| cellspacing="10" style="color:#c0c0c0" | ||
| + | |- | ||
| + | | | ||
| + | {| cellpadding="5" cellspacing="0" style="text-align: center; color:#f3d675; border: 3px solid #4e93a4;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#4e93a4;" | [[Igem2010/Main/synthetic_miR_Kit/July|<font color="#ffecba">July</font>]] | ||
| + | |- style="background:#4e93a4; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- | ||
| + | |colspan="3"| ||'''1'''||'''2'''||'''3'''||'''4''' | ||
| + | |- | ||
| + | |'''5'''||'''6'''||'''7'''||'''8'''||'''9'''||'''10'''||'''11''' | ||
| + | |- | ||
| + | |'''12'''||'''13'''||'''14'''||'''15'''||'''16'''||'''17'''||'''18''' | ||
| + | |- | ||
| + | |'''19'''||'''20'''||'''21'''||'''22'''||'''23'''||'''24'''||'''25''' | ||
| + | |- | ||
| + | |'''26'''||'''27'''||'''28'''||'''29'''||'''30'''||'''31'''|| | ||
| + | |- | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | | | ||
| + | {| cellpadding="5" cellspacing="0" style="text-align: center; color:#f3d675; border: 3px solid #4e93a4;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#4e93a4;" | [[Igem2010/Main/synthetic_miR_Kit/August|<font color="#ffecba">August</font>]] | ||
| + | |- style="background:#4e93a4; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- | ||
| + | |colspan="6"| ||'''1''' | ||
| + | |- | ||
| + | |'''2'''||'''3'''||'''4'''||'''5'''||'''6'''||'''7'''||'''8''' | ||
| + | |- | ||
| + | |'''9'''||'''10'''||'''11'''||'''12'''||'''13'''||'''14'''||'''15''' | ||
| + | |- | ||
| + | |'''16'''||'''17'''||'''18'''||'''19'''||'''20'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#21/08/2010|21]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#22/08/2010|22]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/August#23/08/2010|23]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#21/08/2010|24]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#21/08/2010|25]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#26/08/2010|26]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#27/08/2010|27]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#28/08/2010|28]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#29/08/2010|29]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/August#30/08/2010|30]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#31/08/2010|31]]'''||colspan="5"| | ||
| + | |} | ||
| + | | | ||
| + | {| cellpadding="5" cellspacing="0" style="text-align: center; color:#f3d675; border: 3px solid #4e93a4;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#4e93a4;" | [[Igem2010/Main/synthetic_miR_Kit/September|<font color="#ffecba">September</font>]] | ||
| + | |- style="background:#4e93a4; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- | ||
| + | |colspan="2"| ||'''[[Igem2010/Main/synthetic_miR_Kit/September#01/09/2010|1]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#02/09/2010|2]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#03/09/2010|3]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#04/09/2010|4]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#05/09/2010|5]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/September#06/09/2010|6]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#07/09/2010|7]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#08/09/2010|8]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#09/09/2010|9]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#10/09/2010|10]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#11/09/2010|11]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#12/09/2010|12]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/September#13/09/2010|13]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#14/09/2010|14]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#15/09/2010|15]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#16/09/2010|16]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#17/09/2010|17]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#18/09/2010|18]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#19/09/2010|19]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/September#20/09/2010|20]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#21/09/2010|21]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#22/09/2010|22]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#23/09/2010|23]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#24/09/2010|24]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#25/09/2010|25]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#26/09/2010|26]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/September#27/09/2010|27]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#28/09/2010|28]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#29/09/2010|29]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/September#30/09/2010|30]]'''||colspan="3"| | ||
| + | |- | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | | | ||
| + | {| cellpadding="5" cellspacing="0" style="text-align: center; color:#f3d675; border: 3px solid #4e93a4;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#4e93a4;" | [[Igem2010/Main/synthetic_miR_Kit/October|<font color="#ffecba">October</font>]] | ||
| + | |- style="background:#4e93a4; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- | ||
| + | |colspan="4"| ||'''[[Igem2010/Main/synthetic_miR_Kit/October#01/10/2010|01]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#02/10/2010|02]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#03/10/2010|03]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/October#04/10/2010|04]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#05/10/2010|05]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#06/10/2010|06]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#07/10/2010|07]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#08/10/2010|08]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#09/10/2010|09]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#10/10/2010|10]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/October#11/10/2010|11]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#12/10/2010|12]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#13/10/2010|13]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#14/10/2010|14]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#15/10/2010|15]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#16/10/2010|16]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#17/10/2010|17]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/October#18/10/2010|18]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#19/10/2010|19]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#20/10/2010|20]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#21/10/2010|21]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#22/10/2010|22]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#23/10/2010|23]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#24/10/2010|24]]''' | ||
| + | |- | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/October#25/10/2010|25]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#26/10/2010|26]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/October#27/10/2010|27]]'''||'''28'''||'''29'''||'''30'''||'''31''' | ||
| + | |- | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |} | ||
| + | |||
{{:Team:Heidelberg/Bottom}} | {{:Team:Heidelberg/Bottom}} | ||
Revision as of 02:17, 24 October 2010

|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"