Team:Heidelberg/Notebook/miMeasure
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Laura Nadine (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{:Team:Heidelberg/Template}} | {{:Team:Heidelberg/Template}} | ||
{{:Team:Heidelberg/Pagetop|note_miMeasure}} | {{:Team:Heidelberg/Pagetop|note_miMeasure}} | ||
| - | + | __NOTOC__ | |
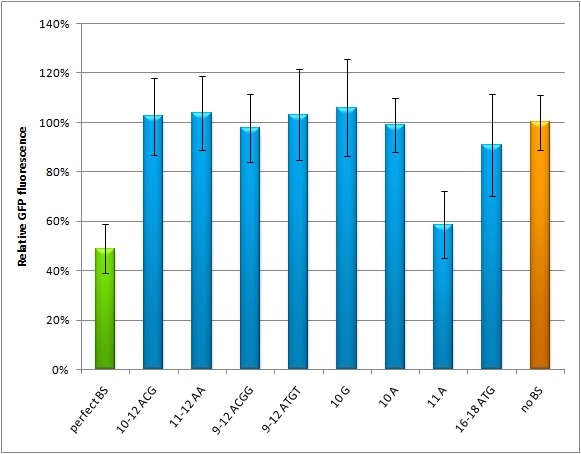
=miMeasure= | =miMeasure= | ||
| Line 103: | Line 103: | ||
|- style="background:#f2f2f2; color:#78b41e" | |- style="background:#f2f2f2; color:#78b41e" | ||
|colspan="7"| | |colspan="7"| | ||
| + | <span style="color:#ffffff">-</span> | ||
|} | |} | ||
Revision as of 00:38, 27 October 2010

| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"