Team:Heidelberg/Notebook/miMeasure
From 2010.igem.org
(Difference between revisions)
(→Seeding and transfection of cells for microscopy and flow cytometry) |
(→Seeding and transfection of cells for microscopy and flow cytometry) |
||
| Line 31: | Line 31: | ||
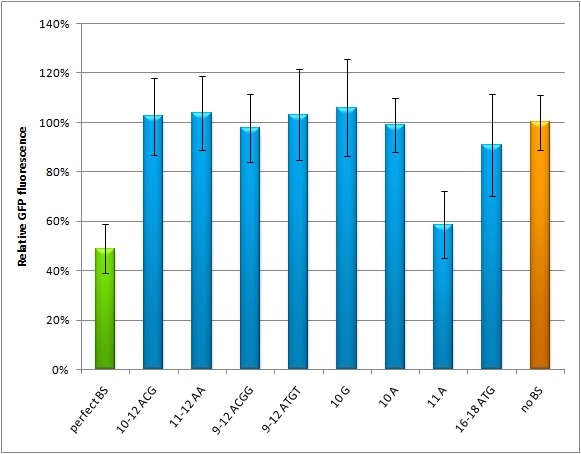
|sequence||binding site feature||Name/number|| | |sequence||binding site feature||Name/number|| | ||
|- | |- | ||
| - | | | + | |ctcagtttactagtgccatttgttc||perfect binding site against miRsAg||M12 |
|- | |- | ||
| - | | | + | |ctcagtttactagacgcatttgttc||miMeasure with randomised nucleotides 10-12||M13 |
|- | |- | ||
| - | | | + | |ctcagtttactagtaacatttgttc||miMeasure with randomised nucleotides 10-12||M14 |
|- | |- | ||
| - | | | + | |ctcagtttactagacggatttgttc||miMeasure with randomised nucleotides 9-12||M15 |
|- | |- | ||
| - | | | + | |ctcagtttactagatgtatttgttc||miMeasure with randomised nucleotides 9-12||M16 |
|- | |- | ||
| - | | | + | |ctcagtttactagtggcatttgttc||miMeasure with mutated nucleotide 10||M17 |
|- | |- | ||
| - | | | + | |ctcagtttactagtgacatttgttc||miMeasure with mutated nucleotide 10||M18 |
|- | |- | ||
| - | | | + | |ctcagtttactagtaccatttgttc||miMeasure with mutated nucleotide 9||M20 |
|- | |- | ||
| - | | | + | |ctcagttatgtagtgccatttgttc||miMeasure with mutated nucleotide 9||M22 |
|- | |- | ||
|-||miMeasure without any binding site||NC (negative control) | |-||miMeasure without any binding site||NC (negative control) | ||
Revision as of 22:02, 26 October 2010

| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"