Team:Heidelberg/Modeling
From 2010.igem.org
| Line 1: | Line 1: | ||
{{:Team:Heidelberg/Single}} | {{:Team:Heidelberg/Single}} | ||
| - | |||
{{:Team:Heidelberg/Single_Pagetop|mode_over}} | {{:Team:Heidelberg/Single_Pagetop|mode_over}} | ||
{{:Team:Heidelberg/Side_Top}} | {{:Team:Heidelberg/Side_Top}} | ||
Revision as of 22:57, 26 October 2010

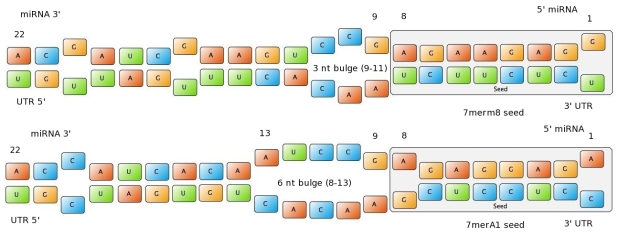
shRNA binding site featuresAs the title of our project states, “DNA is not enough”. There are several upper-level regulation systems in superior organisms. Our main idea was using miRNA to tune down the expression of genes, having tissue-specific, exactly tuned gene therapy as objective. miRNA are non-coding regulatory RNAs functioning as post-transcriptional gene silencers. After they are processed, they are usually 22 nucleotides long and they usually bind to the 3’UTR region of the mRNA (although they can also bind to the ORF or to the 5'UTR), forcing the mRNA into degradation or just repressing translation. In vegetal organisms, miRNA usually bind to the mRNA with extensive complementarity. In animals, interactions are more inexact, creating a lot of uncertainty in the in silico prediction of targets. The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA, and it usually requires extensive pairing. (For the sake of simplicity, we extended slightly the term seed to include the nucleotides 1-8.) Outside the seed, the existence of supplemental pairing (at least 3 contiguous nucleotides and centered in nucleotides 13-16 of the miRNA) stabilizes the bound complex and increases the efficacy of the binding site. Binding sites with a high local AU density around the binding site have proven to be more effective (possibly because of the destabilization of the mRNA secondary structure around the site). The position of the binding site not in the middle of the 3’UTR but either at the end or 15 nt after the stop codon increases the efficacy of repression. IS THE FOLLOWING REALLY NECESSARY? I DON'T THINK I CAN MANAGE TO EVER FIT THE TOPIC HERE... About the mechanistics of the repression, it has been shown that the repressive effect is much higher when the binding site for the miRNA is in the first 15 nt of the 3'UTR. This would match the hypothesis.... BLA BLA BLA Targetscan Scores Jan ???? Figure 1: Interactions between two miRNAs and their binding sites. Examples to show different types of seeds.
Tissue specific miRNAsReferences
|
 "
"