Team:Harvard/allergy/notebook
From 2010.igem.org
(Difference between revisions)
(→08-06-2010 [ top ]) |
(→08-09-2010 [ top ]) |
||
| Line 215: | Line 215: | ||
==08-09-2010 [ [[#top|top]] ]== | ==08-09-2010 [ [[#top|top]] ]== | ||
| + | * DTLs of amiRNA insert were unsuccessful. Yesterday we concluded this was because turbo cells + amp resistance don't grow on YEB media (we were out of LB+amp media). Today we must adopt a new hypothesis as 0/16 colonies grew on LB+amp media. Working theory is that the backbone digestions pictured below have the digested backbone and inserts as the two bands (top to bottom) instead of the undigested & digested backbone (in hindsight, I was trying to ligate into YFP*2, the original insert of B21). The confusion arose from the 1kb ladder, which at first appeared to match another ladder far better than 1kb (see Thursday's entry or thereabouts). This, combined with the fact that the 1kb ladder was hand-labeled, led me to suspect that it was in fact the other ladder (pR322 as pictured [http://openwetware.org/wiki/Image:Ladders.jpg here]). This, in turn, supported the (theorized incorrect) conclusion that the top & bottom bands [http://openwetware.org/wiki/Image:Bbcut1.jpg here] and [http://openwetware.org/wiki/Image:Bbcut2.jpg here] were undigested & digested backbone, respectively, and that the insert had diffused. I've DTL'd again, this time using the upper band (which didn't separate into digested & undigested bands after running for 30m on a .7% agarose gel) for ligation. We'll see on Monday how it went. | ||
| + | *New Plan of action: | ||
| + | #Redigest V9/V10 (three digests of each backbone to use more DNA in ligations) | ||
| + | #Grew up V9/V10+ ihpRNA colonies | ||
| + | #Re pcrd amiRNA and digested to ligate into V9/V10 | ||
| + | #Catalog parts to be ready for the registry | ||
==08-10-2010 [ [[#top|top]] ]== | ==08-10-2010 [ [[#top|top]] ]== | ||
Revision as of 02:52, 24 October 2010

notebook calendar
06-14-2010 [ top ]
06-15-2010 [ top ]
06-16-2010 [ top ]
06-17-2010 [ top ]
06-18-2010 [ top ]
06-21-2010 [ top ]
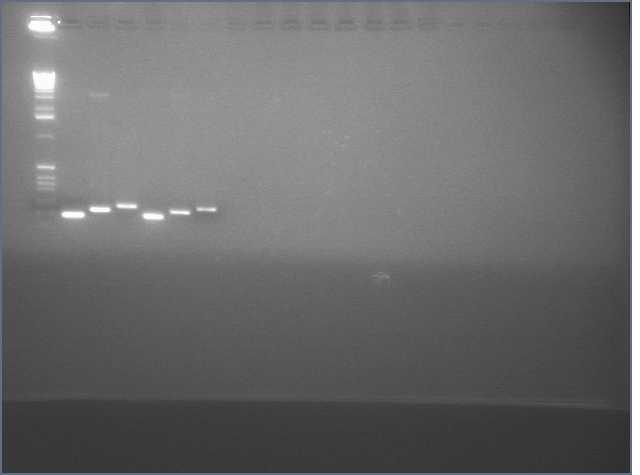
- PCR amplification of gDNA from Arabadopsis thaliana for sense and antisense parts of LTP, Bet v 1, and Ger3.
- Diagnostic Digest of PCR products
Results
Lane 2 is LTP sense, lanes 11 and 12 are Ger 3 sense and antisense.
06-22-2010 [ top ]
- Gel electrophoresis of PCR products
- Gel digest and purification of LTP sense, Ger sense and Ger antisense
- Digest with Xba and Pst
- Purification of parts
Results
Gel purification yielded 2.6, 7.8, and 7.9 ng/μL of LTP sense, Ger sense, and Ger antisense parts. We were able to obtain 1.5, 4.3 and 0.5 ng/μL of digested LTP sense, Ger 3 sense, and Ger 3 antisense, respectively, from the digest purification.
06-23-2010 [ top ]
- Digest and purification of vector V0120
- Ligation of LTP sense, Ger sense, and Ger antisense parts into V0120
Results
Ligations were plated.
06-24-2010 [ top ]
06-25-2010 [ top ]
06-28-2010 [ top ]
06-29-2010 [ top ]
06-30-2010 [ top ]
07-01-2010 [ top ]
07-02-2010 [ top ]
07-05-2010 [ top ]
07-06-2010 [ top ]
07-07-2010 [ top ]
07-08-2010 [ top ]
07-09-2010 [ top ]
07-12-2010 [ top ]
07-13-2010 [ top ]
07-14-2010 [ top ]
07-15-2010 [ top ]
07-16-2010 [ top ]
07-19-2010 [ top ]
07-20-2010 [ top ]
07-21-2010 [ top ]
07-22-2010 [ top ]
07-23-2010 [ top ]
07-26-2010 [ top ]
07-27-2010 [ top ]
07-28-2010 [ top ]
07-29-2010 [ top ]
07-30-2010 [ top ]
08-02-2010 [ top ]
08-03-2010 [ top ]
- Grew up cultures of completed ihpRNA constructs (Bet, LTP, Ger) in pORE expression vector
- amiRNA PCR
- Will look at results of PCR tommorrow
08-04-2010 [ top ]
Tasks
- amiRNA PCR appears to have worked at every Tm we tried:
- Digested V9/V10 to insert our constructs into
- Realized that we hadn't gel purified our ihpRNA inserts
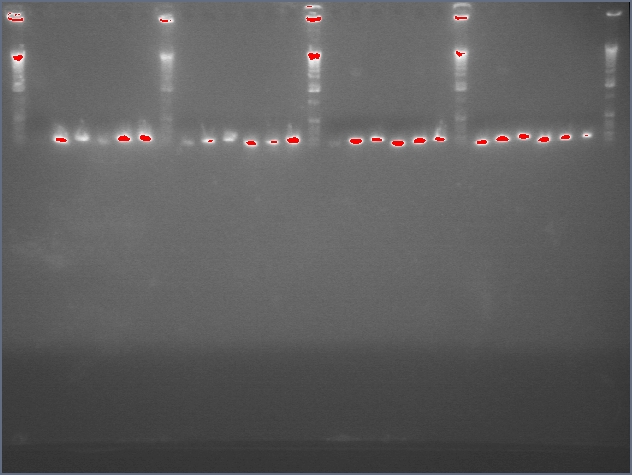
- Gel purification of inserts (entire ihpRNA parts)
Ladder, 9, 11c1, 11c2, ladder, 25c1, 28c1, 28c2, 36c1, 36c2, ladder
Results
- Successfully gel purified our inserts and digested backbones that we will ligate into
08-05-2010 [ top ]
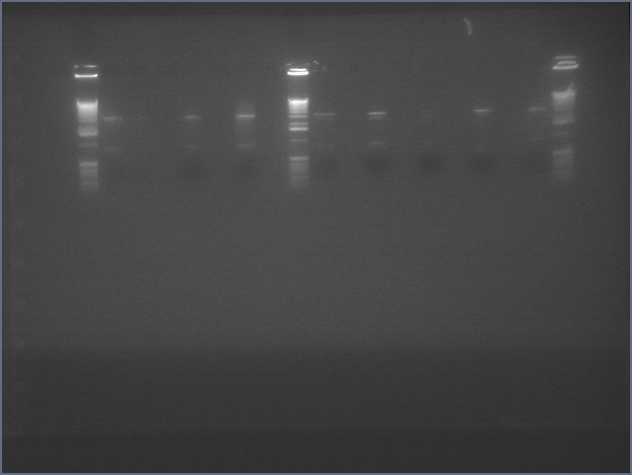
- Gel extracted V9/V10 backbone
Lanes: Ladder, V9, Ladder, V10
Concentrations: V9 (9.4 ng/uL; V10 (16.4 ng/uL)
- Ligated ihpRNA inserts into V9/V10 and transformed
- For our ligations we only used ~ 2uL of backbone (around 18 and 32 ng of backbone)and used a 3x excess of insert
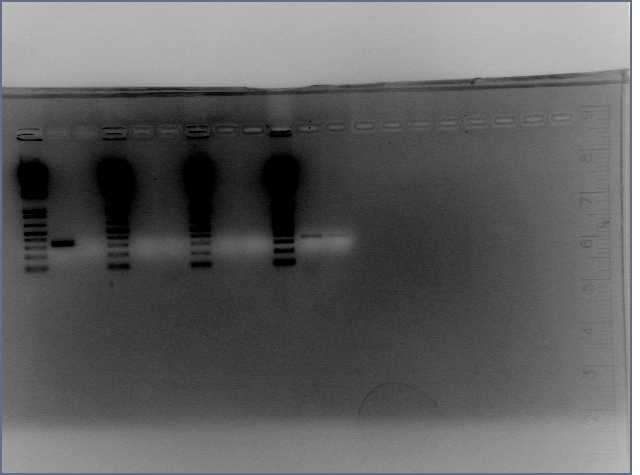
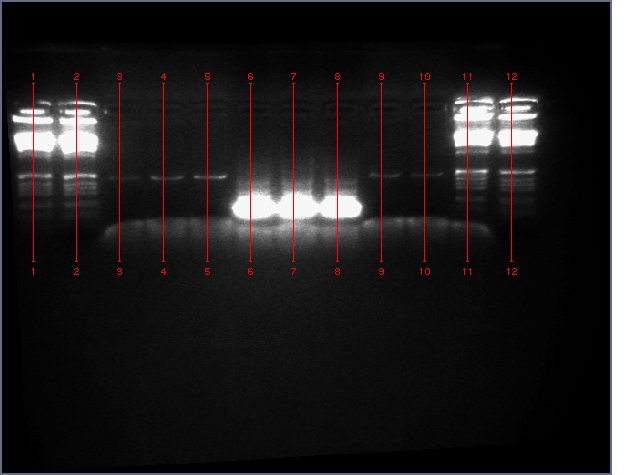
- Verified that amiRNA stitching of Bet, LTP yielded the proper insert with a low level of background through PCR:
Lanes: 1-7: Bet, corresponding to 65.55 degrees C for stitching Tm (even spacing)
Lanes: 8-10: LTP, corresponding to 65.55 degrees C during stitching annealing, even spacing
- Digested
- Bet,LTP inserts with X+P; B21 with X+P+phosphatase
- Ligated, Transformed
08-06-2010 [ top ]
amiRNA
- amiRNA-transformed turbo cells didn't grow in YEB medium. There should be more LB+Amp available by the end of the day. In the meantime, we're digesting some more backbone (8x20ul X+P+B21)
- ligations of Bet/LTP into V0120
ihpRNA
- no colonies on plates from transformations done yesterday after 16 hours (we let these plates grow for longer)
- redoing ligations of ihpRNA inserts in V9/V10
- Redigested and gel purified some backbone
- Concentrations: V9 (5 ng/uL); V10 (6 ng/uL)
- 12 ligations each for V9 and V10
08-09-2010 [ top ]
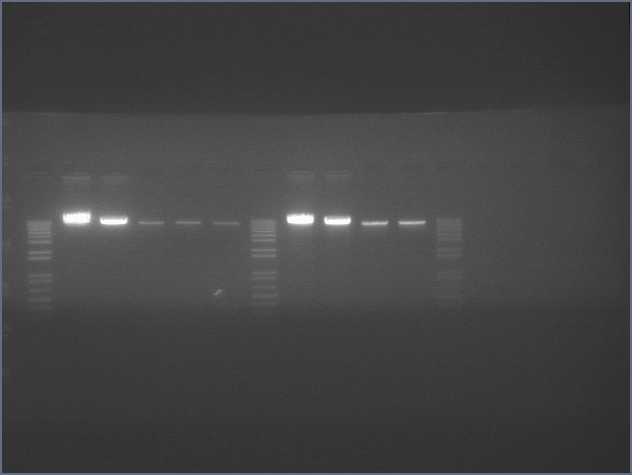
- DTLs of amiRNA insert were unsuccessful. Yesterday we concluded this was because turbo cells + amp resistance don't grow on YEB media (we were out of LB+amp media). Today we must adopt a new hypothesis as 0/16 colonies grew on LB+amp media. Working theory is that the backbone digestions pictured below have the digested backbone and inserts as the two bands (top to bottom) instead of the undigested & digested backbone (in hindsight, I was trying to ligate into YFP*2, the original insert of B21). The confusion arose from the 1kb ladder, which at first appeared to match another ladder far better than 1kb (see Thursday's entry or thereabouts). This, combined with the fact that the 1kb ladder was hand-labeled, led me to suspect that it was in fact the other ladder (pR322 as pictured [http://openwetware.org/wiki/Image:Ladders.jpg here]). This, in turn, supported the (theorized incorrect) conclusion that the top & bottom bands [http://openwetware.org/wiki/Image:Bbcut1.jpg here] and [http://openwetware.org/wiki/Image:Bbcut2.jpg here] were undigested & digested backbone, respectively, and that the insert had diffused. I've DTL'd again, this time using the upper band (which didn't separate into digested & undigested bands after running for 30m on a .7% agarose gel) for ligation. We'll see on Monday how it went.
- New Plan of action:
- Redigest V9/V10 (three digests of each backbone to use more DNA in ligations)
- Grew up V9/V10+ ihpRNA colonies
- Re pcrd amiRNA and digested to ligate into V9/V10
- Catalog parts to be ready for the registry
 "
"