Team:Groningen/9 August 2010
From 2010.igem.org
Week 32
Cell wall isolation streptomyces- Peter
Degredation experiment-Peter
Modellers: Look up and read more articles on the ComXPA quorum sensing system. These pathways can get complicated so after reading a few articles we decided to be old fashioned and draw everything on the blackboard. Joël, Laura, Djoke
Geeske
Transformed E. coli TOP10 with:
-pSB1C3_J04450 (RFP) (plate 1 well 3A 2010 distribution)
-pSB1K3_J04450 (RFP) (plate 1 well 5A 2010 distribution)
These were used to produce pSB1C3_H and pSB1K3_EH via E.coli. Selfclosers would turn red and could easily be ignored, making isolating the backbone from gel unnecessary. Plasmids were checked by restriction.
Arend Jan
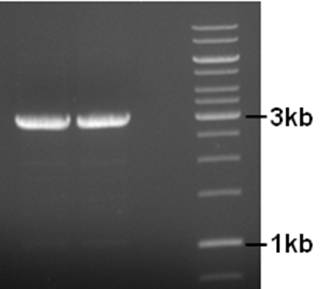
Primers to remove the IS element from the pNZ8901-bbs backbone were designed and ordered. A PCR was performed using the following conditions.
Component µl Final concentration Primer pNZ8901-ins-rem-for 5 300nM Primer pNZ8901-ins-rem-rev 5 300nM 5x Buffer HF 10 1x dNTP’s 2 200µM Template 1 ~1ng Phusion Pol. (Finnzymes) 1 2/1u MQ 26 - 98°C, 3’ - 98°C, 30’’ 25X - 50°C, 30’’ - 72°C, 5’ - 72°C, 10’
Product is of right size. Small amount of by-product (two bands) so annealing temp might be increaed a bit if another PCR is necessary. The used primers have a 5’-ApaI site for circularization. The product was cleaned (BIOKE Nucleospin Extract II kit, great yield), cut with ApaI, cleaned again, ligated to circularize the plasmid, and transformed to coli. Sadly, no transformants. The ligation was repeated, this time with more plasmid. No transformants… getting tired of this. Next week try blunt end ligation of PCR product.
 "
"