Team:Groningen/19 July 2010
From 2010.igem.org
| (21 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
'''Week 29''' | '''Week 29''' | ||
| - | + | ---- | |
<br> | <br> | ||
| Line 9: | Line 9: | ||
<br> | <br> | ||
| - | After lots of meetings the time has come to really start up the modelling part of iGEM. This week we thought about what to model and how. We have looked up and read lots of articles. | + | After lots of meetings the time has come to really start up the modelling part of iGEM. This week we thought about what to model and how. We have looked up and read lots of articles. ''Joël, Arend, Laura, Djoke'' |
<br> | <br> | ||
| Line 37: | Line 37: | ||
<br> | <br> | ||
| - | [[Image:Dispersant1GR.jpg| | + | [[Image:Dispersant1GR.jpg|500px]] |
<br> | <br> | ||
| - | + | Before Shaking | |
<br> | <br> | ||
| - | + | Left to right: Water+oil, Water+oil+CongoRed+AssembledChaplins, Water+Oil+CongoRed+MonomericChaplins, Water+Oil+CongoRed | |
<br> | <br> | ||
| + | |||
| + | [[Image:Dispersant2GR.jpg|500px]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | After Shaking: | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Left to right: Water+oil, Water+oil+CongoRed, Water+Oil+CongoRed+MonomericChaplins, Water+Oil+CongoRed+AssembledChaplins | ||
| + | |||
| + | <br> | ||
| + | |||
| + | As aspected the monomeric chaplins helped to disperse the oil throughout the water, this is nicely visualized by the dispersion of the CongoRed throughout the tube. The assembled chaplins do not show this behavior and seem to have grouped within the oil, which is still on top of the water. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[Image:Dispersant4GR.jpg|300px]] [[Image:Dispersant3GR.jpg|300px]] | ||
| + | <br> | ||
| + | |||
| + | Close ups: Shortly after shaking, left: Control (H20 + Oil), right (Water + Oil + Monomeric Chaplins). | ||
| + | |||
| + | <br> | ||
| + | |||
| + | In close up, it is observed that the monemeric chaplins do indeed help to disperse the oil in the water. As one can see, small droplets of oil are abundant in the water + Chaplins, were no droplets are observed at the same timepoint after shaking in the control. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | '''Arend Jan''' | ||
| + | |||
| + | |||
| + | We’ve transformed all the constructs ordered from Mr. Gene to a new batch of good competent cells and grew single colonies in liquid culture. These were miniprepped and also used to make glycerol stocks. The plasmids were checked by restriction with EcoRI and PstI and had the right size. | ||
| + | |||
| + | |||
| + | It was decided that we will also try to make the pNZ8048 plasmid biobrick compatible so that we can also try to express our chaplins in Lactococcus lactis using this nisin inducible plasmid. The same forward primer can be used to insert the biobricks and another reversed primer was ordered. | ||
| + | |||
| + | |||
| + | <pre>Component µl Final concentration | ||
| + | Primer pNZ89bbs-for1 5 300nM | ||
| + | Primer pNZ8048bbs-rev 5 300nM | ||
| + | 5x Buffer HF 10 1x | ||
| + | dNTP’s 2 200µM | ||
| + | Template 1 ~1ng | ||
| + | Phusion Pol. (Finnzymes)1/0.5 2/1u | ||
| + | MQ 26/26.5 | ||
| + | |||
| + | - 98°C, 3’ | ||
| + | - 98°C, 30’’ | ||
| + | 25X - 50°C, 30’’ | ||
| + | - 72°C, 5’ | ||
| + | - 72°C, 10’ </pre> | ||
| + | |||
| + | |||
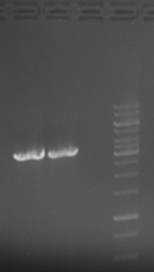
| + | [[Image:23-07-10gn.jpg|200px|thumb|left|Left band = 1ul polymerase, right band = 0.5ul polymerase]] | ||
| + | <br style="clear: both" /> | ||
| + | |||
| + | Products were stored (5ul was loaded on gel) and used for ligation next week. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | '''Geeske''' | ||
| + | |||
| + | ''B. sub'' NZ8900 transformed with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3). Transformation performed according to protocol and transformation-mix was plated on Cm5-plates. Colonies inoculated and glycerol-stocks made of one colony per transformation. | ||
| + | |||
| + | <br> | ||
| + | Chaplin expression tested with only ''B. sub'' NZ8900 pNZ8901_E 6 according to the protocol on the protocol page. Samples were spinned down and supernatant and pellet were stored seperately O/N at -20ºC. Supernatant was put through a TCA-precipitation and subsequently treated with a TFA-treatment (protocol-page). Pellet was put through a TFA-treatment, samples taken for SDS-PAGE. Subsequently the pellets were lysed, dried with a speed-vacuum and then treated with a TFA-treatment. Samples were analyzed with SDS-PAGE by Jorrit, gels colored with silver-staining. Gels were inconclusive (data not shown). | ||
| + | |||
| + | <br> | ||
| + | Transformed ''B. sub'' NZ8900 with genomic DNA from a ''B. sub'' 168 ΔTasA strain according to protocol. Plated on Spec100-plates. Several colonies were inoculated and of 2 glycerol stocks were made. | ||
| + | ''B. sub'' NZ8900 ΔTasA was transformed with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3) according to protocol. First attempt failed. Second attempt finished successfully by Jorrit (described in the next week). | ||
| + | |||
| + | |||
| + | '''Jorrit''' | ||
| + | |||
| + | Finished up the [https://2010.igem.org/Team:Groningen/Protocols_for_Bacillus_subtilis_168 transformation] of Geeske | ||
| + | |||
| + | 200 µL competent NZ8900 cells in five 2 ml reaction vials and added 10 µL of plasmid (2 chpE, 2chpH and negative control) . After one hour at 37 shaker, 200 µL TY was added. After another hour, the vials were plated on Cm5/Km5 TY plates. Colonies were visible the next day. | ||
| + | |||
| + | |||
| + | OD measurement and treating samples according to [https://2010.igem.org/Team:Groningen/Chaplins protocol] | ||
{{Team:Groningen/Footer}} | {{Team:Groningen/Footer}} | ||
Latest revision as of 14:46, 26 October 2010
Week 29
Modellers:
After lots of meetings the time has come to really start up the modelling part of iGEM. This week we thought about what to model and how. We have looked up and read lots of articles. Joël, Arend, Laura, Djoke
David
Drying biofilms
After formation of biofilm on the pieces of wood and ceramics, the material was taken out of the wells plates and put into empty petri dishes. This was put into the 37 C oven for 3 days. Inspection of these dried in biofilms was done by stereomicroscopy.
David & Peter
Dispersant effects chaplin proteins
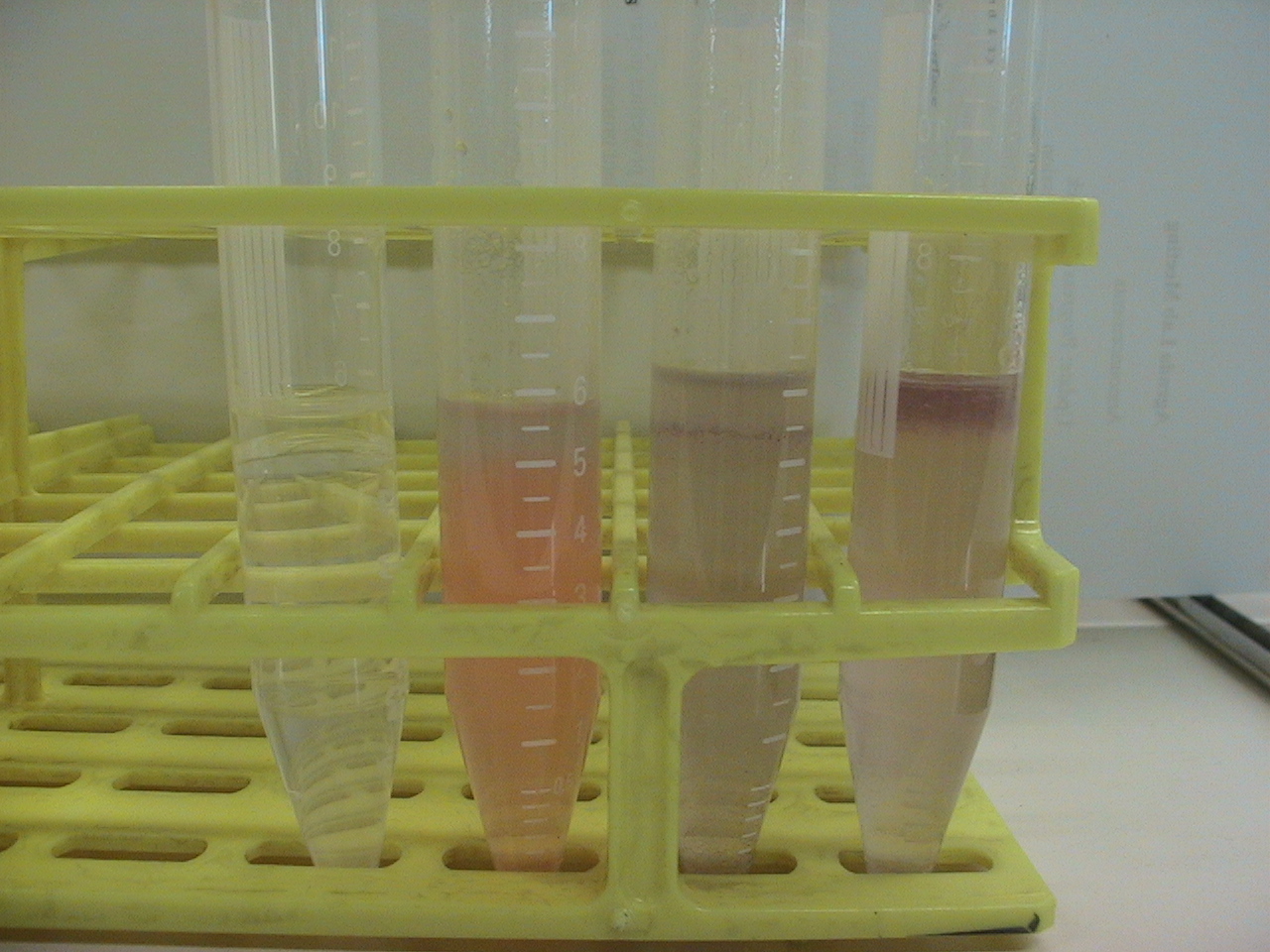
5 ml of demiwater and 0.5 ml olive oil, adding 500 microliter of 15 mg/ml monomeric chaplins to one tube, 0.5 ml vortexed assembled chaplins, and two controls without chpls added. Also ad 30 microliter congo red solution (3 mg/micro liter) to all samples exept one of the two controls. This congo red will stain the assembled chapl Then vortex for 1 min add max speed, then let the samples sit for 10 min. After this closeup pictures are taken of the oil water interface.
Before Shaking
Left to right: Water+oil, Water+oil+CongoRed+AssembledChaplins, Water+Oil+CongoRed+MonomericChaplins, Water+Oil+CongoRed
After Shaking:
Left to right: Water+oil, Water+oil+CongoRed, Water+Oil+CongoRed+MonomericChaplins, Water+Oil+CongoRed+AssembledChaplins
As aspected the monomeric chaplins helped to disperse the oil throughout the water, this is nicely visualized by the dispersion of the CongoRed throughout the tube. The assembled chaplins do not show this behavior and seem to have grouped within the oil, which is still on top of the water.
Close ups: Shortly after shaking, left: Control (H20 + Oil), right (Water + Oil + Monomeric Chaplins).
In close up, it is observed that the monemeric chaplins do indeed help to disperse the oil in the water. As one can see, small droplets of oil are abundant in the water + Chaplins, were no droplets are observed at the same timepoint after shaking in the control.
Arend Jan
We’ve transformed all the constructs ordered from Mr. Gene to a new batch of good competent cells and grew single colonies in liquid culture. These were miniprepped and also used to make glycerol stocks. The plasmids were checked by restriction with EcoRI and PstI and had the right size.
It was decided that we will also try to make the pNZ8048 plasmid biobrick compatible so that we can also try to express our chaplins in Lactococcus lactis using this nisin inducible plasmid. The same forward primer can be used to insert the biobricks and another reversed primer was ordered.
Component µl Final concentration Primer pNZ89bbs-for1 5 300nM Primer pNZ8048bbs-rev 5 300nM 5x Buffer HF 10 1x dNTP’s 2 200µM Template 1 ~1ng Phusion Pol. (Finnzymes)1/0.5 2/1u MQ 26/26.5 - 98°C, 3’ - 98°C, 30’’ 25X - 50°C, 30’’ - 72°C, 5’ - 72°C, 10’
Products were stored (5ul was loaded on gel) and used for ligation next week.
Geeske
B. sub NZ8900 transformed with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3). Transformation performed according to protocol and transformation-mix was plated on Cm5-plates. Colonies inoculated and glycerol-stocks made of one colony per transformation.
Chaplin expression tested with only B. sub NZ8900 pNZ8901_E 6 according to the protocol on the protocol page. Samples were spinned down and supernatant and pellet were stored seperately O/N at -20ºC. Supernatant was put through a TCA-precipitation and subsequently treated with a TFA-treatment (protocol-page). Pellet was put through a TFA-treatment, samples taken for SDS-PAGE. Subsequently the pellets were lysed, dried with a speed-vacuum and then treated with a TFA-treatment. Samples were analyzed with SDS-PAGE by Jorrit, gels colored with silver-staining. Gels were inconclusive (data not shown).
Transformed B. sub NZ8900 with genomic DNA from a B. sub 168 ΔTasA strain according to protocol. Plated on Spec100-plates. Several colonies were inoculated and of 2 glycerol stocks were made.
B. sub NZ8900 ΔTasA was transformed with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3) according to protocol. First attempt failed. Second attempt finished successfully by Jorrit (described in the next week).
Jorrit
Finished up the transformation of Geeske
200 µL competent NZ8900 cells in five 2 ml reaction vials and added 10 µL of plasmid (2 chpE, 2chpH and negative control) . After one hour at 37 shaker, 200 µL TY was added. After another hour, the vials were plated on Cm5/Km5 TY plates. Colonies were visible the next day.
OD measurement and treating samples according to protocol
 "
"