Team:Georgia State/Project
From 2010.igem.org
Zacstewart (Talk | contribs) (→Degradation of Toluene using Genetically Engineered Cyanobacteria) |
(→2010 Georgia State iGEM project) |
||
| (32 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Georgia_State/Header}} | {{Georgia_State/Header}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | |||
| + | |||
| + | |||
| + | ==2010 Georgia State iGEM project== | ||
| + | |||
| + | '''Purpose #1: Tool Kit''' [[Image:Pichia_tool_box_picture.jpg|right|300px]] | ||
| + | |||
| + | The primary purpose of the Georgia State 2010 team is to standardize parts that will facilitate future use of methylotrophic yeast "Pichia pastoris" as a novel chassis for iGEM purposes. We are titling this assembly of Pichia relevant parts as the “Pichia Tool Kit”, which is intended to be a constantly growing and improving resource. So far parts have included promoters, auxotrophic-mutant complementary proteins, selectivity parts and an antigen for intended production. Below is a diagram illustrating the parts that are currently in progress. Click on each image to go directly to the specific tool and get more information. | ||
| + | [[Image:AOX1.png|left|170px]] [[Image:FLD1.png|170px]] [[Image:HA antigen.png|170px]] | ||
| + | |||
| + | |||
| + | [[Image:His4.png|170px]] [[Image:pGAP.png|170px]] [[Image:pTEF.png|170px]] | ||
| + | |||
| + | <code><nowiki>[[Image:His4.png|170px|http://partsregistry.org/Part:BBa_K431006 |alt=Alt text|Title text]]</nowiki></code> | ||
| + | |||
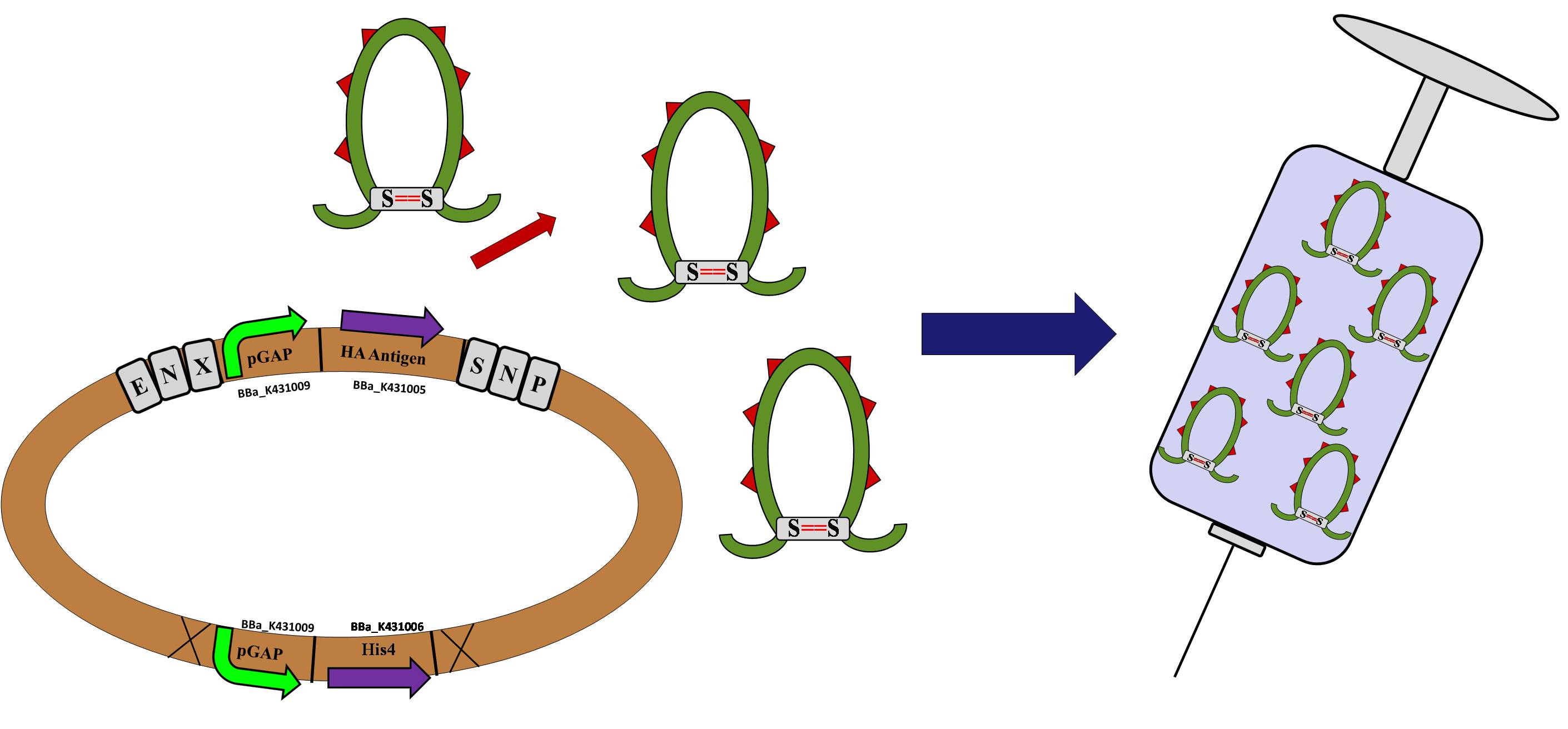
| + | '''Purpose #2: Vaccine Production''' | ||
| + | |||
| + | Because of these reasons, ''P.pastoris'' has quickly gained popularity for recombinant protein production. The Georgia State 2010 team believes ''P. pastoris'' would be an excellent chassis for the iGEM competition. Our goal is to provide a tool box of parts necessary for the genetic manipulation of this organism. Parts will include a plasmid backbone, several parts providing alternative selectivity options and promoter systems. In addition, our tool box will be used to produce a flu virus antigen in ''P. pastoris'' as an example of how this system could be used for vaccine production. We hope our contributions will enable future users to maximize the use and further explore the incredible potential ''P. pastoris'' has to offer! | ||
| + | |||
| + | [[Image:vaccine.jpg|right|700px]] | ||
{{Georgia_State/Footer}} | {{Georgia_State/Footer}} | ||
Latest revision as of 00:50, 28 October 2010

2010 Georgia State iGEM project
Purpose #1: Tool KitThe primary purpose of the Georgia State 2010 team is to standardize parts that will facilitate future use of methylotrophic yeast "Pichia pastoris" as a novel chassis for iGEM purposes. We are titling this assembly of Pichia relevant parts as the “Pichia Tool Kit”, which is intended to be a constantly growing and improving resource. So far parts have included promoters, auxotrophic-mutant complementary proteins, selectivity parts and an antigen for intended production. Below is a diagram illustrating the parts that are currently in progress. Click on each image to go directly to the specific tool and get more information.


[[Image:His4.png|170px|http://partsregistry.org/Part:BBa_K431006 |alt=Alt text|Title text]]
Purpose #2: Vaccine Production
Because of these reasons, P.pastoris has quickly gained popularity for recombinant protein production. The Georgia State 2010 team believes P. pastoris would be an excellent chassis for the iGEM competition. Our goal is to provide a tool box of parts necessary for the genetic manipulation of this organism. Parts will include a plasmid backbone, several parts providing alternative selectivity options and promoter systems. In addition, our tool box will be used to produce a flu virus antigen in P. pastoris as an example of how this system could be used for vaccine production. We hope our contributions will enable future users to maximize the use and further explore the incredible potential P. pastoris has to offer!
 "
"