Team:Georgia State/Aboutthetoolbox
From 2010.igem.org
(→How we made our parts) |
|||
| (3 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | |||
| - | Part Design: Using primers that contain Biobrick ends, parts were isolated from Pichia pastoris chromosomal DNA with PCR reactions. To increase the likelihood of gaining a successful product, a systematic approach was applied for primer design. Two sets of primers were constructed for each part using IDT®’s PrimerQuest and Oligoanalyzer programs. The first set added an Xpa I site on the forward and a Spe I site on the reverse. The second set added EcoRI, Not I and Xba I sites on the forward and | + | |
| + | |||
| + | =='''How we made our parts''' == | ||
| + | |||
| + | |||
| + | |||
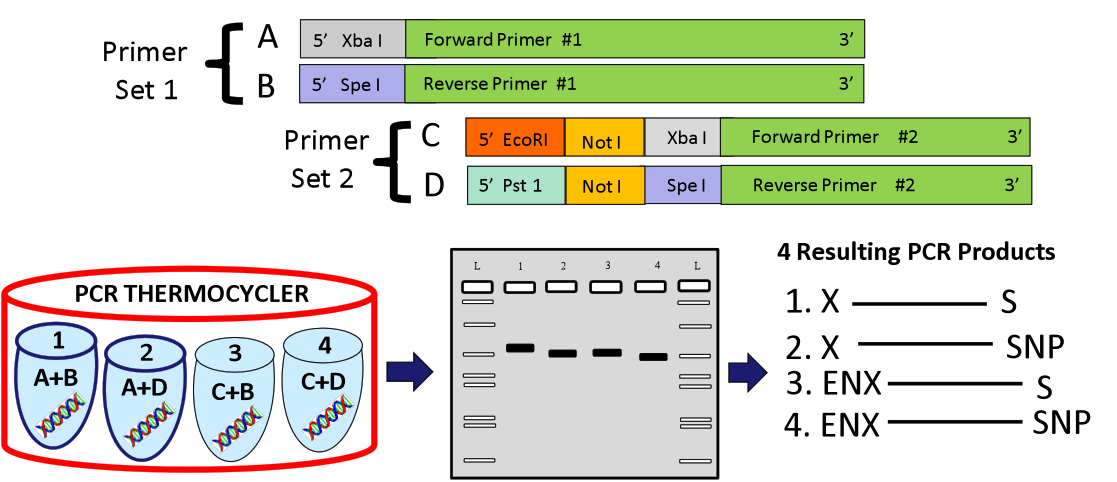
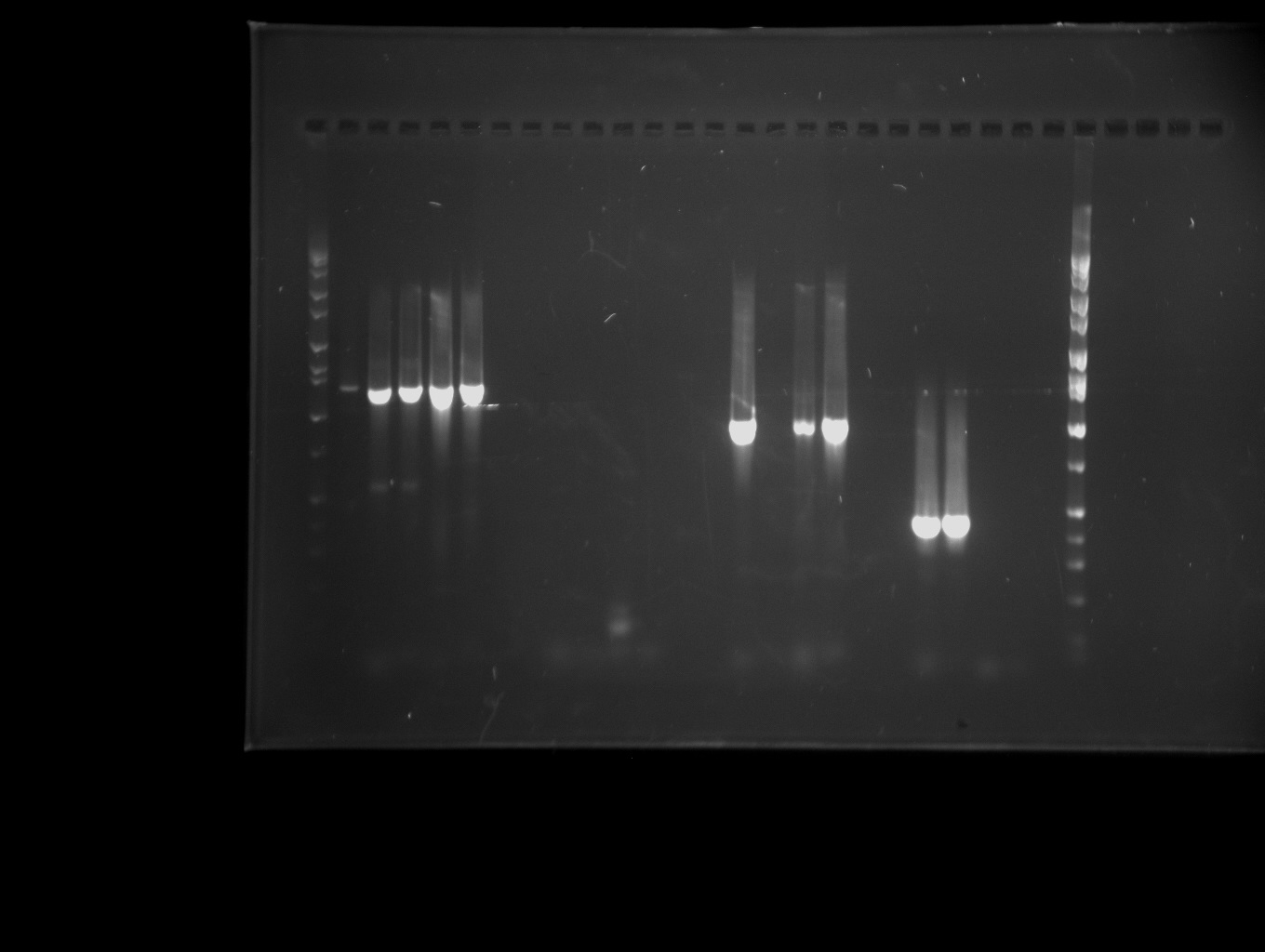
| + | Part Design: Using primers that contain Biobrick ends, parts were isolated from ''Pichia pastoris'' chromosomal DNA with PCR reactions. To increase the likelihood of gaining a successful product, a systematic approach was applied for primer design. Two sets of primers were constructed for each part using ''IDT®’s PrimerQuest'' and ''Oligoanalyzer'' programs. The first set added an Xpa I site on the forward and a Spe I site on the reverse. The second set added EcoRI, Not I and Xba I sites on the forward and Pst I, Not I and Spe I sites on the reverse. In this way, hybrid primer combinations were assembled providing a total of four PCR products. With this system, we have successfully acquired a biobrick prospective product for each part. | ||
| Line 14: | Line 19: | ||
[[Image:iGEMgel.jpg|center|400px]] | [[Image:iGEMgel.jpg|center|400px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
{{Georgia_State/Footer}} | {{Georgia_State/Footer}} | ||
Latest revision as of 03:18, 28 October 2010

How we made our parts
Part Design: Using primers that contain Biobrick ends, parts were isolated from Pichia pastoris chromosomal DNA with PCR reactions. To increase the likelihood of gaining a successful product, a systematic approach was applied for primer design. Two sets of primers were constructed for each part using IDT®’s PrimerQuest and Oligoanalyzer programs. The first set added an Xpa I site on the forward and a Spe I site on the reverse. The second set added EcoRI, Not I and Xba I sites on the forward and Pst I, Not I and Spe I sites on the reverse. In this way, hybrid primer combinations were assembled providing a total of four PCR products. With this system, we have successfully acquired a biobrick prospective product for each part.
 "
"