Team:Freiburg Bioware/testpage
From 2010.igem.org
| (3 intermediate revisions not shown) | |||
| Line 122: | Line 122: | ||
<div class="box_right"> | <div class="box_right"> | ||
</html> | </html> | ||
| - | == | + | ===6. Labday 03.05.2010=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Theoretical cloning</b></p>==== |
| - | + | <p><b>Investigators: Adrian, Hanna, Bea, Patrick, Chris W. </b></p><br> | |
| - | |||
| - | + | There are several matters to do in theoretical cloning. The modularization/modification of the stratagene plasmids and the enzymes. :<br> | |
| - | + | <br> | |
| - | + | 1. Cap-Gen: | |
| - | + | * delete the PstI-restriction site | |
| - | + | * insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick) | |
| + | * add prefix & suffix | ||
| + | * disable binding of Heparan Sulphat Proteoglycan | ||
| + | <br> | ||
| + | 2. Rep-Gen: | ||
| + | * delete EcoRI (2x) and PstI (2x) | ||
| + | <br> | ||
| + | 3. ITRs: | ||
| + | * NotI and PstI (in sequence?) flank the ITRs: its the question if we can/must delete them? | ||
| + | * where exactly starts and ends the sequence (Patrick)? | ||
| + | * it should be checked up if there is a possibility to use all of the three ITRs. | ||
| + | * we blasted the ITR (left) of the MCS vektor (Stratagene). we got a 92% "Coverage" with the AAV2 genom (to 100%). from the alignment (Stratagene ITR with ITR of the AAV2 genom) we got:<br> | ||
| + | [[File:Freiburg10 ITR(left)-Stratagene vs. ITR AAV2.jpg|500x500px|]] | ||
| + | You can see, that the ITR sequence beginns 4 bp after the PstI restriction site. thereupon we did a secundary structure analyse (www.dinamelt.bioinfo.rpi.edu), with the following structurewe got: <br> | ||
| + | [[Media:Freiburg10_AAV2ITR(left)nachBlast.pdf]] <br> | ||
| + | conclusion: the big loop of the secondary structure dosent change if we delete the PstI restriction site (cf. with other uploadet secondary structures), it should be considered that we can't add random bp that could affect the secundary structure. <br> | ||
| + | <b>To do: which bp, which sequence could/can be insertet? cf. the genome of AAV2</b> <br> | ||
| + | <br> | ||
| + | 4. MCS: | ||
| + | * replacement through the iGEM-MCS | ||
| + | * where is the beginning and ending of the MCS in the Vector? ß-globin function and sequence (search for literature and patents, which are denoted in the AAV-Helper-Free-System Manual) | ||
| + | * in this context it would be also important to find out where the beta-Globulin-Intron exactly starts or rather we can cut out the MCS. | ||
| + | <br> | ||
| + | 5. enzymes: | ||
| + | * Thymidinkinase: find informations. TK30 (Bea), SR39 (Hanna) | ||
| + | * Cytosindeaminase: find informations (Adrian) | ||
| + | <br> | ||
| + | in addition we downloadet, addet and anotated the sequence of the pHelper plasmid of stratagene in Geneious (see "Constructs"). | ||
| - | |||
| - | |||
| - | |||
| - | + | insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==== | + | ===7. Labday 07.05.2010=== |
| - | + | ||
| - | + | ||
| - | <p | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Protocolls</b></p>==== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <p><b>Investigators: Anissa, Kerstin </b></p><br> |
| - | + | ||
| - | <br | + | |
| - | + | * adaption and extension of the standart prorcolls (Cloning for Pro's) | |
| - | + | * we startet to create a protokoll-mask for the praktical-cloning (short version for labwork) | |
| - | + | ||
| - | + | ===8. Labday 10.05.2010 === | |
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Stock solutions</b></p>==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ====<p style="font-size:15px; background-color:# | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p><b>Investigators: Adrian, Chris W., Chris L., Bea, Achim, Patrick, Hanna, (Sven)</b></p><br> | |
| + | The following stock solutions were prepared: <br> | ||
| + | 1. Antibiotics: | ||
| + | * Ampicillin: 2 g Ampicillin were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at -20°C. | ||
| + | * Chloramphenicol: 0.5 g Chloramphenicol were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at the -20°C. | ||
| + | * Kanamycin: 1 g Kanamycin was dissolved in 20 mL multipore-H2O, sterilized by filtration and filled into 2 mL tubes and stored at the -20°C. | ||
| + | * Tetracyclin: 0.5 g Tetracyclin were dissolved in 20 mL ethanol (70%) and filled into 2 mL tubes. The tubes were wrapped with aluminium foil (light sensitive!) and stored at -20°C. | ||
| + | 2. ITPG solution (1 M): | ||
| + | * ~ 4.766 g ITPG were dissolved in 20 mL multipore-H2O, sterilized by filtration, filled in 2 mL tubes and stored at -20°C. | ||
| + | 3. DYT (5 litres) | ||
| + | * 80 g Bactotrypton, 50 g Bactoyeast, 25 g NaCl were weight out. | ||
| + | * 2 L multipore-H2O were added | ||
| + | * after mixing, multipore-H2O was added -> endvolume 5 litres | ||
| + | * medium was filled into flask and was autoclaved | ||
| + | 4. Glycerol: | ||
| + | * Glycerol was filled into a flask and was then autoclaved | ||
| - | + | '''To do: register at Mr. Gene!!!''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | ===9. Labday 17.05.2010=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>β-Globin</b></p>==== | |
| - | + | <p><b>Investigators: Bea, Chris W., Patrick, Hanna (and instructors)</b></p><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | We blasted the sequence of the β-globin and additional nukleotides „vorne und hintendran“. | |
| - | + | We found only 70% coverage with the human β-globin-intron add. some parts of the exon 3. | |
| - | + | For this reason we think that the pAAV_MCS annotation of the β-Globin-intron of Stratagene is too generously.<br> | |
| - | + | In addition the alignment of the human ß-globin showed that only parts of the intron 2 (5’) and exon 3 are integratet into the vector. | |
| - | + | We assume that the informations from Stratagene are not total correctly ( intron flanket by the splicedonors and the acceptor-sequence). | |
| - | + | We blasted the sequence between the CMV-promoter and the origin beginning of the ß-globin intron. | |
| + | We found a 98,8% coverage with a synthetic CMV-promoterconstruct ( 1 nucleotide difference). | ||
| + | Literature: „Diverse plasmid DNA vectors by directed molecular evolution of cytomegalovirus promoters. (Wright A. et al.)“<br> | ||
| + | →The question arises if we can omit the ß-globin (because the exact function is unknown). | ||
| + | For this we should contact various Companys (GeneArt, DNA2.0, Mr. Gene,…) to get more information. | ||
| + | In addition we could test the expression with and without ß-globin. | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ===10. Labortag 18.05.2010=== | |
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>pCMV_mVenus_YFP</b></p>==== | |
| - | < | + | |
| - | + | <p><b>Investigator: Bea, Chris W., Patrick, Hanna, Anissa, Kerstin, Adrian (und Instructors)</b></p> | |
| - | + | ||
| - | <b>Investigator: | + | |
| - | < | + | |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | Theoretical cloning with Geneious of pCMV-MCS + pGA14_mVenus_YFP --> <b>pCMV_mVenus_YFP</b> | ||
<ul> | <ul> | ||
| - | <li> | + | <li>Enzyme set: RFC 25 (iGEM) |
| - | <li> | + | <li> digest pGA14_mVenus_YFP (insert) with XbaI and PstI: mVenus_YFP_cut_XbaI+PstI |
| - | <li> | + | <li> digest pCMV_MCS (vector) with XbaI and PstI: pCMV_MCS_cut_XbaI+PstI |
| - | < | + | <li> ligate mVenus_YFP_cut_XbaI+PstI with pCMV_MCS_cut_XbaI+PstI |
| + | <br> | ||
| + | [[File:Freiburg10 pCMV MCS mVenus YFP.png|500x500px|]] | ||
</ul> | </ul> | ||
| - | ==== | + | ===11. Labortag 19.05.2010=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pCMV-MCS + pGA14_mVenus_YFP --> <b>pCMV_mVenus_YFP</b></p>==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p style="font-size:15px; | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <b> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p><b>Investigators: Adrian, Bea, Chris W., Hanna, Patrick</b></p> | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <b>Digestion</b> | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | <ul> | |
| - | < | + | <li>plasmid: insert: pGA14_mVenus_YFP; number: P1 production date: ____ origin: ____ </li> |
| - | + | <li>plasmid: vector: pCMV_MCS; number: P2 production date: ____ origin: ____ </li> | |
| - | + | <li>new vector name: pCMV_mVenus_YFP <br></li> | |
| - | + | <li>buffer used:3 ; Restriction-enzymes used: Enzyme XbaI (no. Lab:___) ; Enzyme PstI(no.Lab:___)</li> | |
| - | + | <li>DNA concentration (vector): 375 ng/µl ; DNA concentration (insert): 476 ng/µl</li> | |
{| border="1" | {| border="1" | ||
| - | | components || align="right" | | + | | components || align="right" | V (pGA_mVenus_YFP)/ µl ||align="right"| V(pCMV_MCS) / µl |
|- | |- | ||
| - | | DNA || align="right" | 2, | + | | DNA || align="right" | 4||align="right"|2,7 |
|- | |- | ||
| - | | BSA ( | + | | BSA (10x) || align="right" |2||align="right"|2 |
|- | |- | ||
| - | | Buffer | + | | Buffer 3 (10x)|| align="right" |2||align="right"|2 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme: XbaI (no.Lab:___)|| align="right" |1,5||align="right"|1,5 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme: PstI (no.Lab:___)|| align="right" |1||align="right"|1 |
|- | |- | ||
| - | | | + | |H2O|| align="right" |9,5||align="right"|10,8 |
|- | |- | ||
| - | |'''Total volume | + | |'''Total volume'''|| align="right" |<b>20</b>||align="right"|<b>20</b> |
|} | |} | ||
| - | <li> Incubation: 1 h | + | <li> Incubation: 1 h at 37°C</li> |
| - | < | + | </ul> |
| - | < | + | |
| - | + | <b>1% Agarose gel and Gel extraction</b> | |
| - | < | + | <ul> |
| - | < | + | <li>prepare 1% agarose gel, run gel for 45 minutes(119 V)</li> |
| + | <li>cut out insert and vector</li> | ||
| + | <li>perform gel extraction following standard protocol provided by Qiagen</li> | ||
| + | </ul> | ||
| - | + | <b>Ligation</b> | |
| - | < | + | |
| - | + | <ul> | |
| - | + | <li>Measure DNA-concentration with Nanodrop </li> | |
| - | < | + | <li>c(mVenus_YFP) = 16,8 ng/µL</li> |
| - | < | + | <li>c(pCMV_MCS) = 22,8 ng/µL</li> |
| - | + | <li>Calculation of volume needed for ligation: | |
| + | <li>c(mVenus_YFP) = 3,66 µL</li> | ||
| + | <li>c(pCMV_MCS) = 5,34 µL</li></li> | ||
| + | </ul> | ||
| - | [[ | + | <b>Transformation</b> |
| - | < | + | <ul> |
| + | <li>Transformation has been followed the standard protocol [[Media:Freiburg10_Cloning Protocol.pdf]] </li> | ||
| + | </ul> | ||
| - | ==== | + | ===12. Labortag 20.05.2010=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Picking clones</b></p>==== |
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | <b> | + | <p><b>Investigators: Adrian, Bea</b></p> |
| - | + | <ul><br/> | |
| - | < | + | Clones were picked according to the standard protocol. |
| + | *3 approaches from each plate</li> | ||
| + | </ul> | ||
| - | ==== | + | ====13. Labortag 21.05.2010: CMV-Promoter==== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | <b>Theoretical cloning:</b> (Volker, Hanna) | |
| - | + | * The CMV promoter (mainly the regulatory region) was further characterized: For this purpose U.S. patent no. 5,385,839 was used. [[Media:Freiburg10_Patent_US5385839A.pdf]] | |
| - | [[File: | + | * Further on the CMV promoter sequence was blasted. The results delivered a 98% query coverage with the "Human herpesvirus 5 strain Toledo, complete genome" (accession no.: GU937742.1). Interestingly the maximal identity was just 99%. This can be explained due to a nucleotide deletion and a C-T transition (red circles), which were also marked in Geneious. |
| - | + | [[File:Freiburg10_CMV-Alignment.jpg|500x500px|]] | |
<br> | <br> | ||
| - | + | [[File:Freiburg10 CMV.jpg|800x800px|]] | |
<br> | <br> | ||
| - | + | '''To do''': find "+1"-location (transcription start); which transcription factors bind to the regulatory region of the CMV promoter? | |
<br> | <br> | ||
| + | <br> | ||
| + | '''LB medium''' was prepared: (Patrick and Chris W.) | ||
| + | * 10 g Bacto-Tryptone, 5 g Bacto-Yeast, 10 g NaCl were mixed in 500 mL milipore-H2O. | ||
| + | * Volume was adjusted to 1 L with milipore-H2O. | ||
| + | * 100 mL flasks were each filled with 50 mL medium. | ||
| + | * 0.75 g agar was added to each flask. | ||
| + | * LB was sterilized by autoclaving and is now stored at room temperature. | ||
| + | <br> | ||
| + | <br> | ||
| + | <b>Plasmid Mini-Prep according to the standard protocol</b> | ||
| - | + | <ul> | |
| - | + | <li>investigator: Adrian, Kira, Anna, Chris W., Patrick</li> | |
| - | + | <li>Measure DNA-concentration with Nanodrop</li> | |
| - | + | </ul> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{| border="1" | {| border="1" | ||
| - | | | + | | || align="right" | P3 (pCMV_mVenus_YFP) ||align="right"| P4 (pCMV_mVenus_YFP) ||align="right"| P5 (pCMV_mVenus_YFP) |
|- | |- | ||
| - | | | + | | concentration (ng/µl)|| align="right" | 457||align="right"|470,8|| align="right" | 477,29 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | ===14. Labortag 25.05.2010=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pCMV_mVenus_YFP + pAAV_MCS</b></p>==== | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | <p><b>Investigators: Kira, Anna, Volker, Jessica</b></p> | |
| - | + | <br> | |
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <li>plasmid: insert: pCMV_mVenus_YFP; number: P5 production date: 21.05.2010 origin: ____ | |
| + | <li>plasmid: vector: pAAV_MCS; number: P? production date: ____ origin: ____ | ||
| + | <li>new vector name: pAAV_mVenus_YFP <br> | ||
| - | + | <br> | |
| - | + | '''1st try: | |
| - | + | <li>buffer used:3 ; Restriction-enzymes used: Enzyme NotI (no. Lab:46) | |
| - | + | <li>DNA concentration (vector): 360 ng/µl ; DNA concentration (insert): 477 ng/µl | |
| - | + | ||
| - | + | ||
| - | <li> | + | |
| - | + | ||
| - | + | ||
| - | <li> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{| border="1" | {| border="1" | ||
| - | | align=" | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pAAV_MCS) / µl |
|- | |- | ||
| - | | align=" | + | | DNA || align="right" | 4.2||align="right"|3.5 |
|- | |- | ||
| - | | align=" | + | | BSA (10x) || align="right" |3||align="right"|3 |
|- | |- | ||
| - | | align=" | + | | Buffer 3 (10x)|| align="right" |3||align="right"|3 |
|- | |- | ||
| - | | align=" | + | |Enzyme: NotI (no.Lab:46)|| align="right" |1||align="right"|1 |
|- | |- | ||
| - | | align=" | + | |H2O|| align="right" |15.3||align="right"|15.3 |
|- | |- | ||
| - | | align=" | + | |'''Total volume'''|| align="right" |<b>26.4</b>||align="right"|<b>25.8</b> |
| - | + | ||
| - | + | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <li> Incubation: 1 1/2 h at 37°C | ||
<br> | <br> | ||
| - | + | '''note: too little water was added | |
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | <br> | |
| - | + | 1% Agarose gel | |
| - | + | <br> | |
| - | + | <li>1% agarose gel was prepared, gel ran for 45 minutes(119 V) | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | '''2nd try:''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <li>buffer used:4 ; Restriction-enzymes used: Enzyme NotI (no. Lab:159) | |
| - | < | + | <li>DNA concentration (vector): 360 ng/µl ; DNA concentration (insert): 477 ng/µl |
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | | components || align="right" | | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pAAV_MCS) / µl |
|- | |- | ||
| - | | DNA || align="right" | | + | | DNA || align="right" | 4.2||align="right"|3.5 |
|- | |- | ||
| - | | | + | | BSA (10x) || align="right" |3||align="right"|3 |
|- | |- | ||
| - | | | + | | Buffer 4 (10x)|| align="right" |3||align="right"|3 |
|- | |- | ||
| - | | | + | |Enzyme: NotI (no.Lab:159)|| align="right" |1,5||align="right"|1,5 |
|- | |- | ||
| - | | | + | |H2O|| align="right" |18,3||align="right"|19 |
|- | |- | ||
| - | |'''Total volume '''|| align="right" | | + | |'''Total volume'''|| align="right" |<b>30</b>||align="right"|<b>30</b> |
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | + | <li> Incubation: 1 1/2 h at 37°C | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | 1% Agarose gel | |
| - | < | + | <br> |
| - | + | <li>1% agarose gel was repared, gel ran for 45 minutes(119 V) | |
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <font color=#FF00FF>Results: unexpected sizes of fragments (see protocol), try again next day with an ethidium bromid gel</font> | ||
<br> | <br> | ||
| - | + | <br> | |
| + | <br> | ||
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Production of chemical competent E.coli</b></p>==== | ||
| - | + | <p><b>Investigators: Patrick and Jessica</b></p> | |
| + | <br> | ||
| + | competent E.coli XL-1-blue and competent E.coli BL21 produced according to the standard-protocoll [[Media:production of competent E.coli.pdf]]<br> | ||
| + | <br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Design of MCS-Oligos</b></p>==== | |
| + | <p><b>Investigators: Bea, Adrian, Hanna, Sven</b></p> | ||
| + | <br> | ||
| + | In order to replace the multiple cloning site of the pAAV_MCS vector from Stratagene by RFC25, two oligos were designed. After their hybridization the overhangs correspond to ClaI- and BglII restriction sites. A digestion with BglII and ClaI will be performed with pAAV-MCS and then will be ligated with the designed oligos.<br> | ||
| + | The oligos (see link) were ordered at Sigma-Aldrich. Estimated shipment: 31.05.2010 .<br> | ||
| - | |||
| - | |||
| - | |||
| - | [[ | + | [[File:Freiburg10 Oligos MCS RFC25 for pAAV.pdf]] |
| - | + | ===15. Labortag 26.05.2010=== | |
| - | < | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Oligos (PstI + MCS)</b></p>==== |
| - | + | ||
| - | + | <p><b>Investigators: Bea, Hanna, Jessica</b></p> | |
| - | < | + | <ul> |
| - | + | <li>Repeat cloning of pAAV-MCS + pCMV_mVenus_YFP --> <b>Goal: pAAV_mVenus_YFP</b><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Cloning did not work (see lab day 25.05.2010) either with NotI or with NotI-HF. Gel did not show any proper band at the expected size. <br> | |
| - | + | There will be used EtBr instead of gelred in the agarose gel and the incubation time of digestion will be prolonged. Further details are followed. <br> | |
| - | + | NOTE: There are three restriction sites of NotI in the pCMV-mVenus_YFP. If cloning with NotI, the polyA hGH signal will be deleted. therefore cloning with NotI is '''not''' possible . | |
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | < | + | <li>Test digestion of pCMV-mVenusYFP with PstI and MluI <br> |
| - | <b>Digestion | + | <b>Digestion</b> |
<br> | <br> | ||
| + | <ul> | ||
| + | <li>experiment date: 26.05.2010 </li> | ||
| + | <li>plasmid: pCMV_mVenus_YFP; number: P5; production date: 21.05.2010/ pCMV-MSC; number: P2; production date: </li> | ||
| + | <li>buffer used: 3/4; Restriction-enzymes used: Enzyme PstI (no. Lab:48) and Enyzme MluI (no. Lab:40) / NotI HF (no. Lab:159) </li> | ||
| + | <li>DNA concentration plasmid: P5 477,3 ng/µl / P2 550ng/µl </li> | ||
| + | |||
{| border="1" | {| border="1" | ||
| - | | components || | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pCMV_MCS) / µl |
|- | |- | ||
| - | | DNA || | + | | DNA || align="right" | 4,2||align="right"|3,6 |
|- | |- | ||
| - | | BSA (10x) || | + | | BSA (10x) || align="right" |2||align="right"|2 |
|- | |- | ||
| - | | Buffer | + | | Buffer 3 (10x)|| align="right" |2||align="right"|2 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme: PstI/NotI HF (no.Lab:48/159)|| align="right" |1||align="right"|2 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme: MluI (no.Lab:40)|| align="right" |1||align="right"|- |
| - | + | ||
|- | |- | ||
| - | |H2O || | + | |H2O|| align="right" |9,8||align="right"|10,4 |
|- | |- | ||
| - | |'''Total volume | + | |'''Total volume'''|| align="right" |<b>20</b>||align="right"|<b>20</b> |
|} | |} | ||
| - | <br | + | <br> |
| + | <li> Incubation: 1 h at 37°C | ||
| - | + | <br> | |
| + | 1% Agarose gel | ||
| + | <br> | ||
| + | <li>1% agarose gel was repared, gel ran for 45 minutes(119 V) | ||
| + | <br> | ||
| + | Results: Test digestion of pCMV_MCS with NotI delivered plausibel results (expected fragment size: ~ 27 kbp and 17 kbp). | ||
| + | Unfortunately the digestion of pCMV_mVenus_YFP with PstI and MluI resulted in one 2.9 kbp and one 1.2 kbp fragment, which didn't correspond to the expected fragment sizes of 19 kbp and 33 kbp - but to fragment sizes of pCMV_MCS without mVenus_YFP! | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <li>'''Ordering oligos:'''(Sigma-Aldrich) <br> | ||
| + | (Bea)<br> | ||
| + | Oligos have been ordered for modifying the ITRs. Deletion of PstI restriction site in ITR. <br> | ||
| - | < | + | <br> |
| - | + | '''right ITR of pAAV_MCS''' <br> | |
| - | + | oligos ordered:<br> | |
| - | + | ||
| + | fwd: 5´- gcgcagctgcctgcaCGGGCGCCTGATGCGG -3´ 77 °C, 31 bp <br> | ||
| + | rev: 5´- CCGCATCAGGCGCCCGTGCAGGCAGCTGCGC -3´ <br> | ||
| + | <br> | ||
| - | + | '''leftITR of pAAV_MCS'''<br> | |
| - | + | oligos ordered:<br> | |
| - | <br | + | |
| + | fwd: 5´- CCTTTTGCTCACATGTCGTGCAGGCAGCTGCGCG -3´ 74 °C, 34 bp <br> | ||
| + | rev: 5´- CGCGCAGCTGCCTGCACGACATGTGAGCAAAAGG -3´<br> | ||
| + | <br> | ||
| + | For further details see link <br> | ||
| - | + | http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/4/44/Freiburg10_Oligos_ITR_mutagenesis_for_pAAV_delete_PstI.pdf | |
| - | + | </li> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | Trafo of | + | '''Test-Trafo of chemical competent E.coli cells''' <br> |
| + | (Hanna) | ||
<br> | <br> | ||
| + | The competent E.coli XL-1-blue and competent E.coli BL21 which were prepared the day before were test-transformed with pUC18 - following the standard protocol. Cells were plated on agar plates containing ampicillin and stored over night at 37°C. | ||
| + | Further on two control plates containing ampicillin or kanamycin were prepared. Non-transformed cells were plated on them and also stored over night at 37°C. | ||
| + | <br> | ||
| + | <br> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | ===16. Labortag 27.05.2010=== | ||
| - | ====<p style="font-size:15px; background-color:# | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cell counting</b></p>==== |
| - | <b>Investigator: | + | <p><b>Investigator: Patrick, Adrian, Chris W. Christian L.</b></p> |
| + | * Meeting | ||
| - | + | cell counting: | |
| - | <br | + | <br> |
| - | + | * BL21 + PUC18 Trafo 54*4 = 216 | |
| - | + | * XL1B + PUC18 Trafo 30*4 = 120 | |
| - | + | digitalization of recipe-cards (Christian L.) | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===17. Labortag 31.05.2010=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>pAAV_RC R585A and R588A transistions</b></p>==== |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | In order to alter the tropism of AAV2 several modifications have to be performed. | |
| - | + | First of all the binding to Heparan Sulfate Proteoglycan has to be disrupted. This can be done by R585A and R588A transistions: | |
| - | + | [[File:Freiburg10 R585A R588A.jpg|800px|]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 12:06, 6 September 2010

Contents |
6. Labday 03.05.2010
Theoretical cloning
Investigators: Adrian, Hanna, Bea, Patrick, Chris W.
There are several matters to do in theoretical cloning. The modularization/modification of the stratagene plasmids and the enzymes. :
1. Cap-Gen:
- delete the PstI-restriction site
- insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick)
- add prefix & suffix
- disable binding of Heparan Sulphat Proteoglycan
2. Rep-Gen:
- delete EcoRI (2x) and PstI (2x)
3. ITRs:
- NotI and PstI (in sequence?) flank the ITRs: its the question if we can/must delete them?
- where exactly starts and ends the sequence (Patrick)?
- it should be checked up if there is a possibility to use all of the three ITRs.
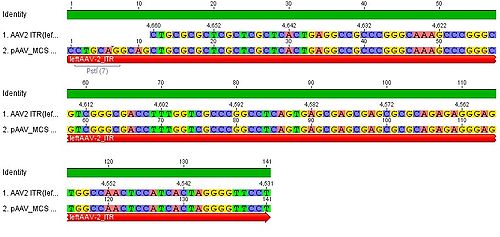
- we blasted the ITR (left) of the MCS vektor (Stratagene). we got a 92% "Coverage" with the AAV2 genom (to 100%). from the alignment (Stratagene ITR with ITR of the AAV2 genom) we got:
You can see, that the ITR sequence beginns 4 bp after the PstI restriction site. thereupon we did a secundary structure analyse (www.dinamelt.bioinfo.rpi.edu), with the following structurewe got:
Media:Freiburg10_AAV2ITR(left)nachBlast.pdf
conclusion: the big loop of the secondary structure dosent change if we delete the PstI restriction site (cf. with other uploadet secondary structures), it should be considered that we can't add random bp that could affect the secundary structure.
To do: which bp, which sequence could/can be insertet? cf. the genome of AAV2
4. MCS:
- replacement through the iGEM-MCS
- where is the beginning and ending of the MCS in the Vector? ß-globin function and sequence (search for literature and patents, which are denoted in the AAV-Helper-Free-System Manual)
- in this context it would be also important to find out where the beta-Globulin-Intron exactly starts or rather we can cut out the MCS.
5. enzymes:
- Thymidinkinase: find informations. TK30 (Bea), SR39 (Hanna)
- Cytosindeaminase: find informations (Adrian)
in addition we downloadet, addet and anotated the sequence of the pHelper plasmid of stratagene in Geneious (see "Constructs").
insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick)
7. Labday 07.05.2010
Protocolls
Investigators: Anissa, Kerstin
- adaption and extension of the standart prorcolls (Cloning for Pro's)
- we startet to create a protokoll-mask for the praktical-cloning (short version for labwork)
8. Labday 10.05.2010
Stock solutions
Investigators: Adrian, Chris W., Chris L., Bea, Achim, Patrick, Hanna, (Sven)
The following stock solutions were prepared:
1. Antibiotics:
- Ampicillin: 2 g Ampicillin were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at -20°C.
- Chloramphenicol: 0.5 g Chloramphenicol were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at the -20°C.
- Kanamycin: 1 g Kanamycin was dissolved in 20 mL multipore-H2O, sterilized by filtration and filled into 2 mL tubes and stored at the -20°C.
- Tetracyclin: 0.5 g Tetracyclin were dissolved in 20 mL ethanol (70%) and filled into 2 mL tubes. The tubes were wrapped with aluminium foil (light sensitive!) and stored at -20°C.
2. ITPG solution (1 M):
- ~ 4.766 g ITPG were dissolved in 20 mL multipore-H2O, sterilized by filtration, filled in 2 mL tubes and stored at -20°C.
3. DYT (5 litres)
- 80 g Bactotrypton, 50 g Bactoyeast, 25 g NaCl were weight out.
- 2 L multipore-H2O were added
- after mixing, multipore-H2O was added -> endvolume 5 litres
- medium was filled into flask and was autoclaved
4. Glycerol:
- Glycerol was filled into a flask and was then autoclaved
To do: register at Mr. Gene!!!
9. Labday 17.05.2010
β-Globin
Investigators: Bea, Chris W., Patrick, Hanna (and instructors)
We blasted the sequence of the β-globin and additional nukleotides „vorne und hintendran“.
We found only 70% coverage with the human β-globin-intron add. some parts of the exon 3.
For this reason we think that the pAAV_MCS annotation of the β-Globin-intron of Stratagene is too generously.
In addition the alignment of the human ß-globin showed that only parts of the intron 2 (5’) and exon 3 are integratet into the vector.
We assume that the informations from Stratagene are not total correctly ( intron flanket by the splicedonors and the acceptor-sequence).
We blasted the sequence between the CMV-promoter and the origin beginning of the ß-globin intron.
We found a 98,8% coverage with a synthetic CMV-promoterconstruct ( 1 nucleotide difference).
Literature: „Diverse plasmid DNA vectors by directed molecular evolution of cytomegalovirus promoters. (Wright A. et al.)“
→The question arises if we can omit the ß-globin (because the exact function is unknown).
For this we should contact various Companys (GeneArt, DNA2.0, Mr. Gene,…) to get more information.
In addition we could test the expression with and without ß-globin.
10. Labortag 18.05.2010
pCMV_mVenus_YFP
Investigator: Bea, Chris W., Patrick, Hanna, Anissa, Kerstin, Adrian (und Instructors)
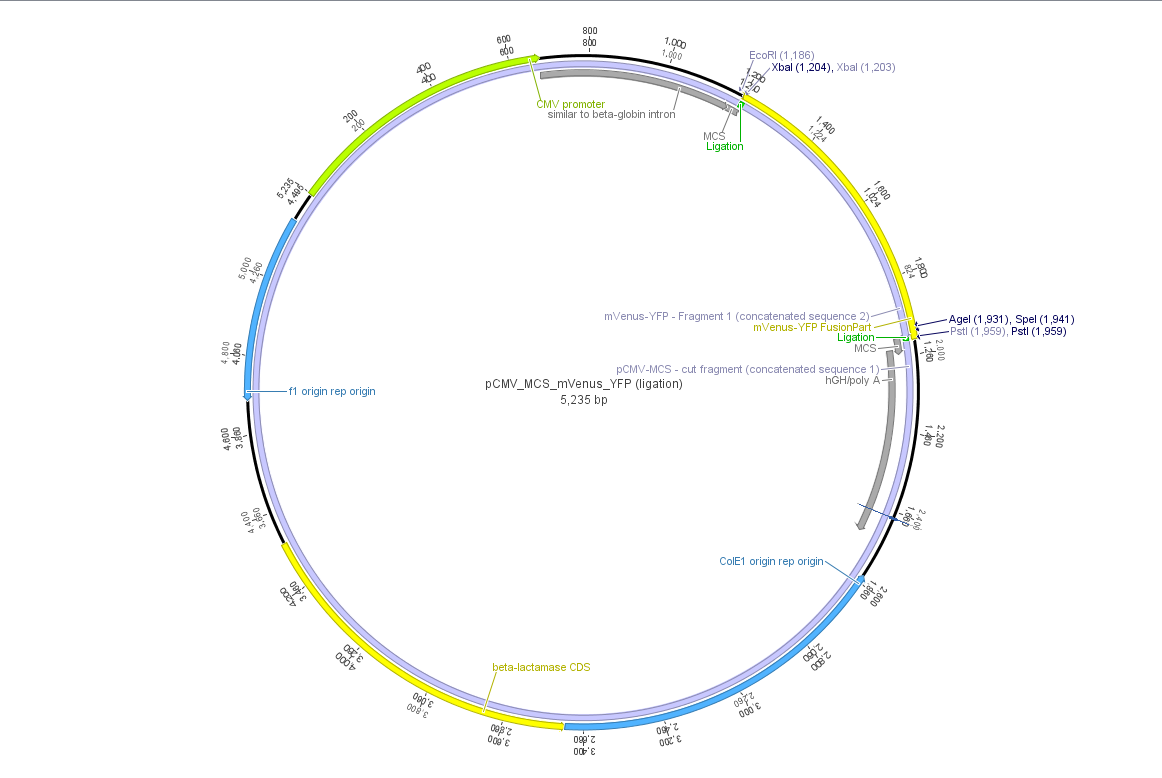
Theoretical cloning with Geneious of pCMV-MCS + pGA14_mVenus_YFP --> pCMV_mVenus_YFP
- Enzyme set: RFC 25 (iGEM)
- digest pGA14_mVenus_YFP (insert) with XbaI and PstI: mVenus_YFP_cut_XbaI+PstI
- digest pCMV_MCS (vector) with XbaI and PstI: pCMV_MCS_cut_XbaI+PstI
- ligate mVenus_YFP_cut_XbaI+PstI with pCMV_MCS_cut_XbaI+PstI
11. Labortag 19.05.2010
Cloning of pCMV-MCS + pGA14_mVenus_YFP --> <b>pCMV_mVenus_YFP
Investigators: Adrian, Bea, Chris W., Hanna, Patrick
Digestion
- plasmid: insert: pGA14_mVenus_YFP; number: P1 production date: ____ origin: ____
- plasmid: vector: pCMV_MCS; number: P2 production date: ____ origin: ____
- new vector name: pCMV_mVenus_YFP
- buffer used:3 ; Restriction-enzymes used: Enzyme XbaI (no. Lab:___) ; Enzyme PstI(no.Lab:___)
- DNA concentration (vector): 375 ng/µl ; DNA concentration (insert): 476 ng/µl
- Incubation: 1 h at 37°C
| components | V (pGA_mVenus_YFP)/ µl | V(pCMV_MCS) / µl |
| DNA | 4 | 2,7 |
| BSA (10x) | 2 | 2 |
| Buffer 3 (10x) | 2 | 2 |
| Enzyme: XbaI (no.Lab:___) | 1,5 | 1,5 |
| Enzyme: PstI (no.Lab:___) | 1 | 1 |
| H2O | 9,5 | 10,8 |
| Total volume | 20 | 20 |
1% Agarose gel and Gel extraction
- prepare 1% agarose gel, run gel for 45 minutes(119 V)
- cut out insert and vector
- perform gel extraction following standard protocol provided by Qiagen
Ligation
- Measure DNA-concentration with Nanodrop
- c(mVenus_YFP) = 16,8 ng/µL
- c(pCMV_MCS) = 22,8 ng/µL
- Calculation of volume needed for ligation:
- c(mVenus_YFP) = 3,66 µL
- c(pCMV_MCS) = 5,34 µL
Transformation
- Transformation has been followed the standard protocol Media:Freiburg10_Cloning Protocol.pdf
12. Labortag 20.05.2010
Picking clones
Investigators: Adrian, Bea
- 3 approaches from each plate
Clones were picked according to the standard protocol.
13. Labortag 21.05.2010: CMV-Promoter
Theoretical cloning: (Volker, Hanna)
- The CMV promoter (mainly the regulatory region) was further characterized: For this purpose U.S. patent no. 5,385,839 was used. Media:Freiburg10_Patent_US5385839A.pdf
- Further on the CMV promoter sequence was blasted. The results delivered a 98% query coverage with the "Human herpesvirus 5 strain Toledo, complete genome" (accession no.: GU937742.1). Interestingly the maximal identity was just 99%. This can be explained due to a nucleotide deletion and a C-T transition (red circles), which were also marked in Geneious.

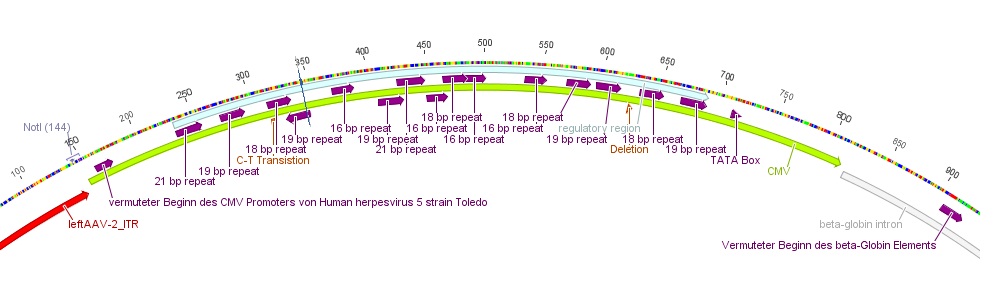
To do: find "+1"-location (transcription start); which transcription factors bind to the regulatory region of the CMV promoter?
LB medium was prepared: (Patrick and Chris W.)
- 10 g Bacto-Tryptone, 5 g Bacto-Yeast, 10 g NaCl were mixed in 500 mL milipore-H2O.
- Volume was adjusted to 1 L with milipore-H2O.
- 100 mL flasks were each filled with 50 mL medium.
- 0.75 g agar was added to each flask.
- LB was sterilized by autoclaving and is now stored at room temperature.
Plasmid Mini-Prep according to the standard protocol
- investigator: Adrian, Kira, Anna, Chris W., Patrick
- Measure DNA-concentration with Nanodrop
| P3 (pCMV_mVenus_YFP) | P4 (pCMV_mVenus_YFP) | P5 (pCMV_mVenus_YFP) | |
| concentration (ng/µl) | 457 | 470,8 | 477,29 |
14. Labortag 25.05.2010
Cloning of pCMV_mVenus_YFP + pAAV_MCS
Investigators: Kira, Anna, Volker, Jessica
1st try:
| components | V (pCMV_mVenus_YFP)/ µl | V(pAAV_MCS) / µl |
| DNA | 4.2 | 3.5 |
| BSA (10x) | 3 | 3 |
| Buffer 3 (10x) | 3 | 3 |
| Enzyme: NotI (no.Lab:46) | 1 | 1 |
| H2O | 15.3 | 15.3 |
| Total volume | 26.4 | 25.8 |
note: too little water was added
1% Agarose gel
2nd try:
| components | V (pCMV_mVenus_YFP)/ µl | V(pAAV_MCS) / µl |
| DNA | 4.2 | 3.5 |
| BSA (10x) | 3 | 3 |
| Buffer 4 (10x) | 3 | 3 |
| Enzyme: NotI (no.Lab:159) | 1,5 | 1,5 |
| H2O | 18,3 | 19 |
| Total volume | 30 | 30 |
1% Agarose gel
Results: unexpected sizes of fragments (see protocol), try again next day with an ethidium bromid gel
Production of chemical competent E.coli
Investigators: Patrick and Jessica
competent E.coli XL-1-blue and competent E.coli BL21 produced according to the standard-protocoll Media:production of competent E.coli.pdf
Design of MCS-Oligos
Investigators: Bea, Adrian, Hanna, Sven
In order to replace the multiple cloning site of the pAAV_MCS vector from Stratagene by RFC25, two oligos were designed. After their hybridization the overhangs correspond to ClaI- and BglII restriction sites. A digestion with BglII and ClaI will be performed with pAAV-MCS and then will be ligated with the designed oligos.
The oligos (see link) were ordered at Sigma-Aldrich. Estimated shipment: 31.05.2010 .
File:Freiburg10 Oligos MCS RFC25 for pAAV.pdf
15. Labortag 26.05.2010
Oligos (PstI + MCS)
Investigators: Bea, Hanna, Jessica
- Repeat cloning of pAAV-MCS + pCMV_mVenus_YFP --> Goal: pAAV_mVenus_YFP
Cloning did not work (see lab day 25.05.2010) either with NotI or with NotI-HF. Gel did not show any proper band at the expected size.
There will be used EtBr instead of gelred in the agarose gel and the incubation time of digestion will be prolonged. Further details are followed.
NOTE: There are three restriction sites of NotI in the pCMV-mVenus_YFP. If cloning with NotI, the polyA hGH signal will be deleted. therefore cloning with NotI is not possible .
- Test digestion of pCMV-mVenusYFP with PstI and MluI
Digestion
- experiment date: 26.05.2010
- plasmid: pCMV_mVenus_YFP; number: P5; production date: 21.05.2010/ pCMV-MSC; number: P2; production date:
- buffer used: 3/4; Restriction-enzymes used: Enzyme PstI (no. Lab:48) and Enyzme MluI (no. Lab:40) / NotI HF (no. Lab:159)
- DNA concentration plasmid: P5 477,3 ng/µl / P2 550ng/µl
- Incubation: 1 h at 37°C
1% Agarose gel
- 1% agarose gel was repared, gel ran for 45 minutes(119 V)
Results: Test digestion of pCMV_MCS with NotI delivered plausibel results (expected fragment size: ~ 27 kbp and 17 kbp). Unfortunately the digestion of pCMV_mVenus_YFP with PstI and MluI resulted in one 2.9 kbp and one 1.2 kbp fragment, which didn't correspond to the expected fragment sizes of 19 kbp and 33 kbp - but to fragment sizes of pCMV_MCS without mVenus_YFP!
- Ordering oligos:(Sigma-Aldrich)
(Bea)
Oligos have been ordered for modifying the ITRs. Deletion of PstI restriction site in ITR.
right ITR of pAAV_MCS
oligos ordered:
fwd: 5´- gcgcagctgcctgcaCGGGCGCCTGATGCGG -3´ 77 °C, 31 bp
rev: 5´- CCGCATCAGGCGCCCGTGCAGGCAGCTGCGC -3´
leftITR of pAAV_MCS
oligos ordered:
fwd: 5´- CCTTTTGCTCACATGTCGTGCAGGCAGCTGCGCG -3´ 74 °C, 34 bp
rev: 5´- CGCGCAGCTGCCTGCACGACATGTGAGCAAAAGG -3´
For further details see link
http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/4/44/Freiburg10_Oligos_ITR_mutagenesis_for_pAAV_delete_PstI.pdf
components V (pCMV_mVenus_YFP)/ µl V(pCMV_MCS) / µl DNA 4,2 3,6 BSA (10x) 2 2 Buffer 3 (10x) 2 2 Enzyme: PstI/NotI HF (no.Lab:48/159) 1 2 Enzyme: MluI (no.Lab:40) 1 - H2O 9,8 10,4 Total volume 20 20
Test-Trafo of chemical competent E.coli cells
(Hanna)
The competent E.coli XL-1-blue and competent E.coli BL21 which were prepared the day before were test-transformed with pUC18 - following the standard protocol. Cells were plated on agar plates containing ampicillin and stored over night at 37°C. Further on two control plates containing ampicillin or kanamycin were prepared. Non-transformed cells were plated on them and also stored over night at 37°C.
16. Labortag 27.05.2010
Cell counting
Investigator: Patrick, Adrian, Chris W. Christian L.
- Meeting
cell counting:
- BL21 + PUC18 Trafo 54*4 = 216
- XL1B + PUC18 Trafo 30*4 = 120
digitalization of recipe-cards (Christian L.)
17. Labortag 31.05.2010
pAAV_RC R585A and R588A transistions
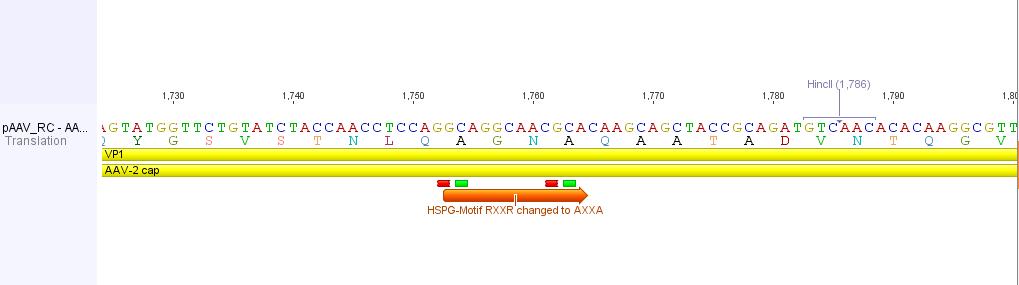
In order to alter the tropism of AAV2 several modifications have to be performed.
First of all the binding to Heparan Sulfate Proteoglycan has to be disrupted. This can be done by R585A and R588A transistions:
 "
"