|
|
| Line 122: |
Line 122: |
| | <div class="box_right"> | | <div class="box_right"> |
| | </html> | | </html> |
| - | ===99. labday 24.08.2010=== | + | ====6. Labday 03.05.2010==== |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of pSB1C3_CFP_middlelinker</b></p>==== | + | |
| - | '''Investigator: Jessica'''
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Theoretical cloning</b></p>==== |
| | + | |
| | + | <p><b>Investigators: Adrian, Hanna, Bea, Patrick, Chris W. </b></p><br> |
| | + | |
| | + | |
| | + | There are several matters to do in theoretical cloning. The modularization/modification of the stratagene plasmids and the enzymes. :<br> |
| | <br> | | <br> |
| | + | 1. Cap-Gen: |
| | + | * delete the PstI-restriction site |
| | + | * insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick) |
| | + | * add prefix & suffix |
| | + | * disable binding of Heparan Sulphat Proteoglycan |
| | <br> | | <br> |
| | + | 2. Rep-Gen: |
| | + | * delete EcoRI (2x) and PstI (2x) |
| | <br> | | <br> |
| - | | + | 3. ITRs: |
| - | [[File:Freiburg10 pSB1C3_CFP_middlelinker.jpg|900px]] | + | * NotI and PstI (in sequence?) flank the ITRs: its the question if we can/must delete them? |
| | + | * where exactly starts and ends the sequence (Patrick)? |
| | + | * it should be checked up if there is a possibility to use all of the three ITRs. |
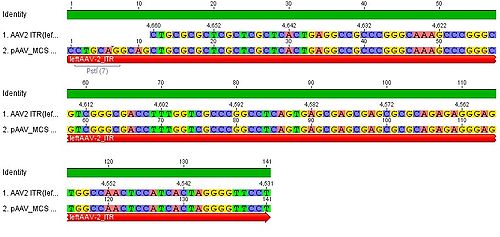
| | + | * we blasted the ITR (left) of the MCS vektor (Stratagene). we got a 92% "Coverage" with the AAV2 genom (to 100%). from the alignment (Stratagene ITR with ITR of the AAV2 genom) we got:<br> |
| | + | [[File:Freiburg10 ITR(left)-Stratagene vs. ITR AAV2.jpg|500x500px|]] |
| | + | You can see, that the ITR sequence beginns 4 bp after the PstI restriction site. thereupon we did a secundary structure analyse (www.dinamelt.bioinfo.rpi.edu), with the following structurewe got: <br> |
| | + | [[Media:Freiburg10_AAV2ITR(left)nachBlast.pdf]] <br> |
| | + | conclusion: the big loop of the secondary structure dosent change if we delete the PstI restriction site (cf. with other uploadet secondary structures), it should be considered that we can't add random bp that could affect the secundary structure. <br> |
| | + | <b>To do: which bp, which sequence could/can be insertet? cf. the genome of AAV2</b> <br> |
| | <br> | | <br> |
| - | * '''pSB1C3_CFP_middlelinker is ready''' | + | 4. MCS: |
| | + | * replacement through the iGEM-MCS |
| | + | * where is the beginning and ending of the MCS in the Vector? ß-globin function and sequence (search for literature and patents, which are denoted in the AAV-Helper-Free-System Manual) |
| | + | * in this context it would be also important to find out where the beta-Globulin-Intron exactly starts or rather we can cut out the MCS. |
| | + | <br> |
| | + | 5. enzymes: |
| | + | * Thymidinkinase: find informations. TK30 (Bea), SR39 (Hanna) |
| | + | * Cytosindeaminase: find informations (Adrian) |
| | + | <br> |
| | + | in addition we downloadet, addet and anotated the sequence of the pHelper plasmid of stratagene in Geneious (see "Constructs"). |
| | | | |
| | | | |
| | + | insert the antibody fragment (targeting): sequence? in which part of VP1? (Patrick) |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Harvest viral particles, Transduction of HT1080</b></p>==== | + | ====7. Labday 07.05.2010==== |
| - | '''Investigator: Kerstin'''
| + | |
| | | | |
| - | *Harvest viral particles following the standard protocol
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Protocolls</b></p>==== |
| | | | |
| - | *Transduction of three 6-well plates:
| + | <p><b>Investigators: Anissa, Kerstin </b></p><br> |
| | | | |
| - | Plate 1 YFP: 150.000 cells per well<br />
| + | * adaption and extension of the standart prorcolls (Cloning for Pro's) |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | * we startet to create a protokoll-mask for the praktical-cloning (short version for labwork) |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| | | | |
| - | </tr>
| + | ====8. Labday 10.05.2010 ==== |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>500µl virus (1) </td>
| + | |
| - | <td>500µl virus (2) </td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>500µl virus (1) </td>
| + | |
| - | <td>500µl virus (2) </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | Plate 2 YFP: 150.000 cells per well<br />
| + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| | | | |
| - | </tr>
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Stock solutions</b></p>==== |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>500µl virus (3) </td>
| + | |
| - | <td>500µl virus (4) </td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>500µl virus (3) </td>
| + | |
| - | <td>500µl virus (4) </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | Plate 3 YFP: 200.000 cells per well<br />
| + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr> | + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| | | | |
| - | </tr> | + | <p><b>Investigators: Adrian, Chris W., Chris L., Bea, Achim, Patrick, Hanna, (Sven)</b></p><br> |
| - | <tr> | + | The following stock solutions were prepared: <br> |
| - | <td>A</td> | + | 1. Antibiotics: |
| - | <td>control, no cells</td> | + | * Ampicillin: 2 g Ampicillin were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at -20°C. |
| - | <td>500µl virus (5) </td>
| + | * Chloramphenicol: 0.5 g Chloramphenicol were dissolved in 20 mL ethanol (70%), filled into 2 mL tubes and stored at the -20°C. |
| - | <td>500µl virus (9) </td>
| + | * Kanamycin: 1 g Kanamycin was dissolved in 20 mL multipore-H2O, sterilized by filtration and filled into 2 mL tubes and stored at the -20°C. |
| - | </tr>
| + | * Tetracyclin: 0.5 g Tetracyclin were dissolved in 20 mL ethanol (70%) and filled into 2 mL tubes. The tubes were wrapped with aluminium foil (light sensitive!) and stored at -20°C. |
| - | <tr>
| + | 2. ITPG solution (1 M): |
| - | <td>B</td>
| + | * ~ 4.766 g ITPG were dissolved in 20 mL multipore-H2O, sterilized by filtration, filled in 2 mL tubes and stored at -20°C. |
| - | <td>control, no virus</td>
| + | 3. DYT (5 litres) |
| - | <td> 500µl virus (9) </td>
| + | * 80 g Bactotrypton, 50 g Bactoyeast, 25 g NaCl were weight out. |
| - | <td> 500µl virus (5) </td>
| + | * 2 L multipore-H2O were added |
| - | </tr>
| + | * after mixing, multipore-H2O was added -> endvolume 5 litres |
| - | </table>
| + | * medium was filled into flask and was autoclaved |
| | + | 4. Glycerol: |
| | + | * Glycerol was filled into a flask and was then autoclaved |
| | | | |
| | + | '''To do: register at Mr. Gene!!!''' |
| | + | <br> |
| | | | |
| - | *(1)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,06
| + | ====9. Labday 17.05.2010==== |
| - | *(2)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,08
| + | |
| - | *(3)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,10
| + | |
| - | *(4)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,12
| + | |
| - | *(5)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,14
| + | |
| - | *(9)= 10µg RC, 10µg pHelper, 3,3µg GOI (<b>eGFP</b>)
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-prep of SDM NgoMIV CD</b></p>==== | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>β-Globin</b></p>==== |
| - | '''Investigator: Kira''' <br />
| + | |
| - | Motivation: Harvest DNA for further analysis, e.g. test digestion
| + | |
| | | | |
| - | Mini-prep was performed on 3 colonies.
| + | <p><b>Investigators: Bea, Chris W., Patrick, Hanna (and instructors)</b></p><br> |
| | + | <br> |
| | + | We blasted the sequence of the β-globin and additional nukleotides „vorne und hintendran“. |
| | + | We found only 70% coverage with the human β-globin-intron add. some parts of the exon 3. |
| | + | For this reason we think that the pAAV_MCS annotation of the β-Globin-intron of Stratagene is too generously.<br> |
| | + | In addition the alignment of the human ß-globin showed that only parts of the intron 2 (5’) and exon 3 are integratet into the vector. |
| | + | We assume that the informations from Stratagene are not total correctly ( intron flanket by the splicedonors and the acceptor-sequence). |
| | + | We blasted the sequence between the CMV-promoter and the origin beginning of the ß-globin intron. |
| | + | We found a 98,8% coverage with a synthetic CMV-promoterconstruct ( 1 nucleotide difference). |
| | + | Literature: „Diverse plasmid DNA vectors by directed molecular evolution of cytomegalovirus promoters. (Wright A. et al.)“<br> |
| | + | →The question arises if we can omit the ß-globin (because the exact function is unknown). |
| | + | For this we should contact various Companys (GeneArt, DNA2.0, Mr. Gene,…) to get more information. |
| | + | In addition we could test the expression with and without ß-globin. |
| | | | |
| - | c(clone1) = 358, 05 ng/ul
| + | <br> |
| - | c(clone2) = 352, 10 ng/ul
| + | |
| - | c(clone 3) = 376, 53 ng/ul
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sent for sequencing: pSB1C3_6xHis</b></p>==== | + | ===10. Labortag 18.05.2010=== |
| - | '''Investigator: Jessica'''
| + | |
| - | * Plasmid: '''P84''' c= 107,8 ng/µl
| + | |
| - | * Primer: VR2
| + | |
| - | * tube number: JG84
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Subcloning Cap into pAAV_RC_ins-rep</b></p>==== | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>pCMV_mVenus_YFP</b></p>==== |
| - | '''Investigator: Stefan'''
| + | |
| - | <br />
| + | |
| | | | |
| - | Comment: Sequencing of pAAV_RC_final showed that Cap was not inserted succsessfully. Therefore it has to be repeated.
| |
| - | <br />
| |
| - | <br />
| |
| - | '''Digestion of vector and insert:'''
| |
| - | *Insert: pMA_RepCap Vector_SDM_InsPvuII clone 1 (P211)
| |
| - | *Vector: pAAV_RC_ins-rep clone1 (P250)<br />
| |
| - | <br />
| |
| - | <br />
| |
| - | A two-step digestion was performed: <br />
| |
| - | <br />
| |
| - | '''1st step'''
| |
| - | {| border="1"
| |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| |
| - | |-
| |
| - | |DNA|| align="right" |5,8 || align="right" | 5,2
| |
| - | |-
| |
| - | |buffer 3|| align="right" |2 || align="right" | 2
| |
| - | |-
| |
| - | | Enzyme BsiWI|| align="right" |1 || align="right" | 1
| |
| - | |-
| |
| - | |H<sub>2</sub>O|| align="right" |11,2 || align="right" | 11,8
| |
| - | |-
| |
| - | |total volume|| align="right" |20 || align="right" |20
| |
| - | |}
| |
| - | <br />
| |
| - | '''2nd step'''
| |
| - | {| border="1"
| |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| |
| - | |-
| |
| - | |Mix obtained from step 1 || align="right" |20 || align="right" | 20
| |
| - | |-
| |
| - | | Enzyme BspMI|| align="right" |1 || align="right" | 1
| |
| - | |-
| |
| - | |H<sub>2</sub>O|| align="right" |1,25 || align="right" | 1,25
| |
| - | |-
| |
| - | |total volume|| align="right" |22,25 || align="right" |22,25
| |
| - | |}
| |
| | | | |
| | + | <p><b>Investigator: Bea, Chris W., Patrick, Hanna, Anissa, Kerstin, Adrian (und Instructors)</b></p> |
| | | | |
| - | Samples were loaded on a 0,8% agarose gel an run at 115V for about 60 minutes.
| |
| | | | |
| | + | Theoretical cloning with Geneious of pCMV-MCS + pGA14_mVenus_YFP --> <b>pCMV_mVenus_YFP</b> |
| | + | <ul> |
| | + | <li>Enzyme set: RFC 25 (iGEM) |
| | + | <li> digest pGA14_mVenus_YFP (insert) with XbaI and PstI: mVenus_YFP_cut_XbaI+PstI |
| | + | <li> digest pCMV_MCS (vector) with XbaI and PstI: pCMV_MCS_cut_XbaI+PstI |
| | + | <li> ligate mVenus_YFP_cut_XbaI+PstI with pCMV_MCS_cut_XbaI+PstI |
| | + | <br> |
| | + | [[File:Freiburg10 pCMV MCS mVenus YFP.png|500x500px|]] |
| | + | </ul> |
| | | | |
| - | <gallery widths=400px heights=400px caption="pAAV RC final">
| + | ===11. Labortag 19.05.2010=== |
| - | Image:Freiburg10 Cap Insertion.png
| + | |
| - | </gallery>
| + | |
| | | | |
| | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pCMV-MCS + pGA14_mVenus_YFP --> <b>pCMV_mVenus_YFP</b></p>==== |
| | | | |
| - | '''Gelextraction:'''<br />
| + | <p><b>Investigators: Adrian, Bea, Chris W., Hanna, Patrick</b></p> |
| | | | |
| - | Gelex was performed according to protocol exept for elution was done with 60 µl instead of 20 µl.<br />
| + | <b>Digestion</b> |
| - | *c(P211)= 1,14 ng/µl
| + | <br> |
| - | *c(P250)= 5,60 ng/µl
| + | <ul> |
| - | <br /> | + | <li>plasmid: insert: pGA14_mVenus_YFP; number: P1 production date: ____ origin: ____ </li> |
| - | <br /> | + | <li>plasmid: vector: pCMV_MCS; number: P2 production date: ____ origin: ____ </li> |
| - | '''T4 Ligation:'''<br />
| + | <li>new vector name: pCMV_mVenus_YFP <br></li> |
| - | T4 ligation was performed according to protocol.
| + | <li>buffer used:3 ; Restriction-enzymes used: Enzyme XbaI (no. Lab:___) ; Enzyme PstI(no.Lab:___)</li> |
| - | used DNA amounts: <br />
| + | <li>DNA concentration (vector): 375 ng/µl ; DNA concentration (insert): 476 ng/µl</li> |
| - | *v(P211)= 2,56 µl
| + | |
| - | *v(P250)= 5,44 µl
| + | |
| - | <br /> | + | |
| - | <br /> | + | |
| - | '''Transformation:'''<br />
| + | |
| - | <br /> | + | |
| - | Trafo was performed according to protocol using XL1b cells.
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Test digestion of pCerulean (p273) </b></p>====
| + | |
| - | '''Investigator: Anissa'''<br>
| + | |
| | | | |
| - | <p style="font-size:13px; color:#66bbff;"><b>Comments:</b>Sequencing of pCerulean showed strange results, because the qualtity of sequenzing was not good. For working further with pCerulean, we started with a test digestion, to be sure, everything works.<br/>
| |
| - | Because the sequencing showed no AgeI, we cut one time with AgeI and EcoRI, the other time with NotI.
| |
| - | </p>
| |
| - | <br />
| |
| | {| border="1" | | {| border="1" |
| - | | align="left" | '''Components''' ||align="left"| '''p273/µL''' ||align="left"| '''p273/µL''' | + | | components || align="right" | V (pGA_mVenus_YFP)/ µl ||align="right"| V(pCMV_MCS) / µl |
| | |- | | |- |
| - | | align="left" | DNA ||align="left"| 2||align="left"| 2
| + | | DNA || align="right" | 4||align="right"|2,7 |
| | |- | | |- |
| - | | align="left" | BSA (10x) ||align="left"|1,5||align="left"|1,5
| + | | BSA (10x) || align="right" |2||align="right"|2 |
| | |- | | |- |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1,5||align="left"| 1,5
| + | | Buffer 3 (10x)|| align="right" |2||align="right"|2 |
| | |- | | |- |
| - | | align="left" | Enzyme 1 ||align="left"|0,5 NotI||align="left"|0,5 EcoRI
| + | |Enzyme: XbaI (no.Lab:___)|| align="right" |1,5||align="right"|1,5 |
| | |- | | |- |
| - | | align="left" | Enzyme 2 ||align="left"|0,5 NotI ||align="left"|0,5 AgeI
| + | |Enzyme: PstI (no.Lab:___)|| align="right" |1||align="right"|1 |
| | |- | | |- |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 9||align="left"| 9 | + | |H2O|| align="right" |9,5||align="right"|10,8 |
| | |- | | |- |
| - | | align="left" | '''Total volume''' ||align="left"| 15||align="left"|15
| + | |'''Total volume'''|| align="right" |<b>20</b>||align="right"|<b>20</b> |
| | |} | | |} |
| - | <br />
| |
| - | [[Image:Freiburg10 pCerulean test digestion.jpg|200px|test digestion of pCerulean]]
| |
| - | <br />
| |
| - | <br />
| |
| | | | |
| - | <p style="font-size:13px; color:#66bbff;"><b>Comments:</b> Results of digestion seem to be all right</p>
| + | <li> Incubation: 1 h at 37°C</li> |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of VP1up into PCerulean </b></p>====
| + | |
| - | '''Investigator: Anissa'''<br>
| + | |
| - | VP1up was cut out of pSB1C3 and into pCerulean
| + | |
| - | <ul>
| + | |
| - | <li> Digestion: | + | |
| - | <br /><br />
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| '''Vector/µL''' ||align="left"| '''Insert/µL'''
| + | |
| - | |-
| + | |
| - | | align="left" | DNA ||align="left"| 3,7||align="left"|8,4
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"|1,5||align="left"|2
| + | |
| - | |-
| + | |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1,5||align="left"| 2
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 1 EcoRI ||align="left"|1||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 2 PstI ||align="left"| 1||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | H<sub>2</sub>O ||align="left"|6,3||align="left"| 5,6
| + | |
| - | |-
| + | |
| - | | align="left" | '''Total volume''' ||align="left"|15||align="left"| 20
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | </li> | + | |
| - | <li> Gel:
| + | |
| - | <br/>
| + | |
| - | A 1% agarose-gel was made, after 30 minutes samples were cut<br/>
| + | |
| - | [[Image:Freiburg10 Anissa24810.png|400px|test digestion of pCerulean]]
| + | |
| - | <br/>
| + | |
| - | </li>
| + | |
| - | <li> Gelextraction was performed according the standardprotocol </li>
| + | |
| - | <li> Ligation:
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| '''used volume for T4 ligation/µL''' ||align="left"| '''concentration /ng/µl'''
| + | |
| - | |-
| + | |
| - | | align="left" | Vector ||align="left"| 5,15||align="left"|27,7
| + | |
| - | |-
| + | |
| - | | align="left" |Insert ||align="left"|2,85||align="left"|15,4
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | In addition 1µl T4-ligase and 1µl T4-ligase-buffer were added
| + | |
| - | </li>
| + | |
| - | <li>Transformation: was performes into Xl1 blue according the standard-protocol
| + | |
| - | </li>
| + | |
| - | <br />
| + | |
| | </ul> | | </ul> |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of sequencing reads prepared yesterday (23.08.2010)</b></p>====
| |
| - | <b>Investigator: Bea </b>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#cc0033;"><I><b>GENERAL COMMENT:</b> pSB1C3 {NLS and VP1up} were sent for sequencing. Aswell as the pAAV_RC_final which contains all four mutations in the Rep-Cap gene and the integrated and synthesiezd rep and cap gene and the plasmid pCerulean whith the CMV promoter and sv40 polyadenylation site.</i> </p>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pAAV_RC_final containing the subcloned "rep" (ordered) and "Cap (ordered) plus the four mutations to delete the restriction sites PstI, BamHi and SalI. </p>
| |
| - | <ul>
| |
| - | <li>Primer used: </li>
| |
| - | <ul>
| |
| - | <li>VP1 primer for pKex</li>
| |
| - | <li>GATC_std_SK</li>
| |
| - | <li>Primer Cap 2800 rev</li>
| |
| - | <li>Primer Cap 2800 for</li>
| |
| - | <li>Primer Cap 3500 for</li>
| |
| - | </ul>
| |
| - | <li>Plasmid sequenced: P283 </li>
| |
| - | <li>Sequence sample: iGEM4</li>
| |
| - | <li>Stored in Geneious-folder: RepCap insert ordered</li>
| |
| - | </ul>
| |
| - |
| |
| - | <gallery widths=400px heights=400px caption="pAAV RC final">
| |
| - | Image:Freiburg10 Seqanalysis pAAV RC final 2010 23 08.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | <b>Results:</b> <p style="color:#00bbff;">Sequencing of the mutation ok. Rep integration worked. Cap integration needs to be repeated.</p>
| |
| - |
| |
| - | <br />
| |
| - | <b>Next steps: Repetition of the integration of the synthesized cap gene performed by Stefan (see topic). </b>
| |
| - | <br />
| |
| - | <br />
| |
| | | | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pSB1C3_NLS </p>
| + | <b>1% Agarose gel and Gel extraction</b> |
| | <ul> | | <ul> |
| - | <li>Primer used: VR-2</li> | + | <li>prepare 1% agarose gel, run gel for 45 minutes(119 V)</li> |
| - | <li>Plasmid sequenced: P282 </li> | + | <li>cut out insert and vector</li> |
| - | <li>Sequence sample: iGEM3_O51_VR-2 </li> | + | <li>perform gel extraction following standard protocol provided by Qiagen</li> |
| - | <li>Stored in folder: pSB1C3_NLS</li>
| + | |
| | </ul> | | </ul> |
| | | | |
| - | <gallery widths=600px heights=400px caption="pSB1C3_NLS">
| + | <b>Ligation</b> |
| - | Image: Freiburg10 Sequence analysis pSB1C3 NLS 2010 23 08.jpg
| + | |
| - | </gallery>
| + | |
| - | <br />
| + | |
| - | <b>Results:</b> <p style="color:#00bbff;">Insertion of the nuclear localisation signal (NLS) into the pSB1C3_NLS as one step in the N-terminal insertion of targeting molecules. Sequencing results look good. prefix and suffix ok!</p> | + | |
| | | | |
| - | <br />
| |
| - | <b>Next steps: Fuse NLS to targeting molecule. </b>
| |
| - | <br />
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pSB1C3_VP1up</p>
| |
| | <ul> | | <ul> |
| - | <li>Primer used: VF-2</li> | + | <li>Measure DNA-concentration with Nanodrop </li> |
| - | <li>Plasmid sequenced: P280 </li> | + | <li>c(mVenus_YFP) = 16,8 ng/µL</li> |
| - | <li>Sequence sample: iGEM1_VF-2 </li> | + | <li>c(pCMV_MCS) = 22,8 ng/µL</li> |
| - | <li>Stored in folder: N-Terminal targeting --> pSB1C3_VP1up</li> | + | <li>Calculation of volume needed for ligation: |
| | + | <li>c(mVenus_YFP) = 3,66 µL</li> |
| | + | <li>c(pCMV_MCS) = 5,34 µL</li></li> |
| | </ul> | | </ul> |
| - | <gallery widths=600px heights=400px caption="pSB1C3_VP1up">
| |
| - | Image:Freiburg10 Seqanalysis pSB1C3 VP1up 2010 23 08.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | <b>Results:</b> <p style="color:#00bbff;"> Prefix ok, but annotation is wrong. VP1up should start with: GC!! Suffix ok! The additional EcoRI before the prefix standard can be due to the bad sequence read quality.</p>
| |
| | | | |
| - | <br />
| + | <b>Transformation</b> |
| - | <b>Next steps: Clone VP1up into pCerulean in order to obtain the plasmid with a CMV promoter and the sv40 polyadenylation site. </b> | + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pCerulean </p>
| + | |
| | <ul> | | <ul> |
| - | <li>Primer used: GATC_CMV-F</li> | + | <li>Transformation has been followed the standard protocol [[Media:Freiburg10_Cloning Protocol.pdf]] </li> |
| - | <li>Plasmid sequenced: P273 </li>
| + | |
| - | <li>Sequence sample: iGEM2_CMV-F </li>
| + | |
| - | <li>Stored in folder: N-Terminal targeting --> pCerulean</li>
| + | |
| | </ul> | | </ul> |
| - | <gallery widths=600px heights=400px caption="pCerulean">
| |
| - | Image:Freiburg10 Seqanalysis pCerulean 2010 23 08.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | <b>Results:</b> <p style="color:#00bbff;"> It seem that the suffix is not correct which means that AgeI, PstI and NotI is missing. This can be due to the bad sequence quality. Prefix is ok!
| |
| - | </p>
| |
| | | | |
| - | <br />
| + | ===12. Labortag 20.05.2010=== |
| - | <b>Next steps: For verification a test digestion with the missing restriction sites will be perforemd and another round of sequencing will be conducted. </b>
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| | | | |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Picking clones</b></p>==== |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Test digestion of SDM NgoMIV CD </b></p>====
| + | <p><b>Investigators: Adrian, Bea</b></p> |
| - | '''Investigator: Kira'''<br>
| + | <ul><br/> |
| - | Motivation: In order to check if site directed mutagenesis was successfull, test digestion was performed with 3 clones, which were picked yesterday as well as with 'original' DNA, which still contains all the restriction sites
| + | Clones were picked according to the standard protocol. |
| | + | *3 approaches from each plate</li> |
| | + | </ul> |
| | | | |
| - | <li> Test digestion:
| + | ====13. Labortag 21.05.2010: CMV-Promoter==== |
| - | <br /><br />
| + | <br> |
| - | {| border="1"
| + | <b>Theoretical cloning:</b> (Volker, Hanna) |
| - | | align="left" | '''Components''' ||align="left"| '''clone 1/µL''' ||align="left"| '''clone 2/µL''' ||align="left"| '''clone 3/µL'''||align="left"| '''original/µL'''
| + | * The CMV promoter (mainly the regulatory region) was further characterized: For this purpose U.S. patent no. 5,385,839 was used. [[Media:Freiburg10_Patent_US5385839A.pdf]] |
| - | |-
| + | * Further on the CMV promoter sequence was blasted. The results delivered a 98% query coverage with the "Human herpesvirus 5 strain Toledo, complete genome" (accession no.: GU937742.1). Interestingly the maximal identity was just 99%. This can be explained due to a nucleotide deletion and a C-T transition (red circles), which were also marked in Geneious. |
| - | | align="left" | DNA (500ng) ||align="left"| 1,4||align="left"|1,4||align="left"| 1,4||align="left"| 1,4
| + | [[File:Freiburg10_CMV-Alignment.jpg|500x500px|]] |
| - | |-
| + | <br> |
| - | | align="left" | BSA (10x) ||align="left"| 1||align="left"| 1||align="left"| 1||align="left"| 1
| + | [[File:Freiburg10 CMV.jpg|800x800px|]] |
| - | |-
| + | <br> |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1||align="left"| 1||align="left"| 1||align="left"| 1
| + | '''To do''': find "+1"-location (transcription start); which transcription factors bind to the regulatory region of the CMV promoter? |
| - | |-
| + | <br> |
| - | | align="left" | Enzyme 1 NgoMIV ||align="left"|0,5||align="left"| 0,5||align="left"|0,5||align="left"|0,5
| + | <br> |
| - | |-
| + | '''LB medium''' was prepared: (Patrick and Chris W.) |
| - | | align="left" | H<sub>2</sub>O ||align="left"|6,1||align="left"| 6,1||align="left"| 6,1||align="left"| 6,1
| + | * 10 g Bacto-Tryptone, 5 g Bacto-Yeast, 10 g NaCl were mixed in 500 mL milipore-H2O. |
| - | |-
| + | * Volume was adjusted to 1 L with milipore-H2O. |
| - | | align="left" | '''Total volume''' ||align="left"|10||align="left"| 10||align="left"| 10||align="left"| 10
| + | * 100 mL flasks were each filled with 50 mL medium. |
| - | |}
| + | * 0.75 g agar was added to each flask. |
| - | <br />
| + | * LB was sterilized by autoclaving and is now stored at room temperature. |
| - |
| + | <br> |
| - | <br />
| + | <br> |
| - | [[image:Freiburg10_Kopfkratzen.gif|thumb|right| Bitte schneid mich aus !]] | + | <b>Plasmid Mini-Prep according to the standard protocol</b> |
| - | | + | |
| - | [[file:Freiburg10 test digestion NgoMIV.jpg]] | + | |
| - | | + | |
| - | According to the gel, NgoMIV SDM was successful in clones 1 and 2. Clone 3 well reveales 2 bands of unknown origin. Samples 1 and 3 were sent for sequencing.
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Picking clones of pSB1C3_Affibody_linker and pSB1C3_ß-Globin_YFP_hGH_rITR </b></p>====
| + | |
| - | <b>Investigator: Anna </b>
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: A 1:1000 dilution of pSB1C3_Affibody_GSAT-Linker was prepared. <br/>
| + | |
| - | <b>To do</b>: Mini-Prep of pSB1C3_Affibody_(Short/Middle/Long and SEG-Linker) and pSB1C3_ß-Globin_YFP_hGH_rITR. Picking clones of pSB1C3_Affibody_GSAT-Linker. </p>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Test digestion of pSB1C3_SDM_SspI_Bla14FM (P222), pSB1C3_BLA (P286, P288) </b></p>====
| + | |
| - | <b>Investigator: Achim</b>
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>New test digestion of two SDM-attempts, this time we also digested the original vector containing the PvuII restriction site to see differences.</p>
| + | |
| - | <br/> | + | |
| - | | + | |
| - | | + | |
| - | [[File:Freiburg10 Test digestion of pSB1C3 BLA final 24 08 2010.jpg|800px]]
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | *The original vector is being cut as expected: the uncut vector can be seen in different conformations, the linearized vector shows one distinct band and the digestions cutting out BLA show a band at ~900 bp. Our SDM attempts show inconsistent results, not only does the Pvu site still seem to be in the backbone, there are no more proper insert bands visible either. We therefore conclude that either the DpnI digestion or the dilution of the transformation must have gone wrong (differing plasmids...). Tomorow we'll try a new cloning approach using an old vector with deleted Pvu restriction site and MscI & XbaI digestion. In case this also fails we also yet have to prep and test digest the repetition of the mutagenesis which was carried out by Stefan. | + | |
| - | | + | |
| - | | + | |
| - | </ul> | + | |
| - | <gallery widths=600px heights=400px caption="pSB1C3_VP1up">
| + | |
| - | Image:Freiburg10 Seqanalysis pSB1C3 VP1up 2010 23 08.jpg
| + | |
| - | </gallery>
| + | |
| - | <br /> | + | |
| - | <b>Results:</b> <p style="color:#00bbff;"> Prefix ok, but annotation is wrong. VP1up should start with: GC!! Suffix ok! The additional EcoRI before the prefix standard can be due to the bad sequence read quality.</p> | + | |
| | | | |
| - | <br />
| |
| - | <b>Next steps: Clone VP1up into pCerulean in order to obtain the plasmid with a CMV promoter and the sv40 polyadenylation site. </b>
| |
| - | <br />
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pCerulean </p>
| |
| | <ul> | | <ul> |
| - | <li>Primer used: GATC_CMV-F</li> | + | <li>investigator: Adrian, Kira, Anna, Chris W., Patrick</li> |
| - | <li>Plasmid sequenced: P273 </li> | + | <li>Measure DNA-concentration with Nanodrop</li> |
| - | <li>Sequence sample: iGEM2_CMV-F </li>
| + | |
| - | <li>Stored in folder: N-Terminal targeting --> pCerulean</li>
| + | |
| | </ul> | | </ul> |
| - | <gallery widths=600px heights=400px caption="pCerulean">
| |
| - | Image:Freiburg10 Seqanalysis pCerulean 2010 23 08.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | <b>Results:</b> <p style="color:#00bbff;"> It seem that the suffix is not correct which means that AgeI, PstI and NotI is missing. This can be due to the bad sequence quality. Prefix is ok!
| |
| - | </p>
| |
| | | | |
| - | <br />
| |
| - | <b>Next steps: For verification a test digestion with the missing restriction sites will be perforemd and another round of sequencing will be conducted. </b>
| |
| - | <br />
| |
| - | <br />
| |
| - |
| |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Test digestion of SDM NgoMIV CD </b></p>====
| |
| - | '''Investigator: Kira'''<br>
| |
| - | Motivation: In order to check if site directed mutagenesis was successfull, test digestion was performed with 3 clones, which were picked yesterday as well as with 'original' DNA, which still contains all the restriction sites
| |
| - |
| |
| - | <li> Test digestion:
| |
| - | <br /><br />
| |
| | {| border="1" | | {| border="1" |
| - | | align="left" | '''Components''' ||align="left"| '''clone 1/µL''' ||align="left"| '''clone 2/µL''' ||align="left"| '''clone 3/µL'''||align="left"| '''original/µL''' | + | | || align="right" | P3 (pCMV_mVenus_YFP) ||align="right"| P4 (pCMV_mVenus_YFP) ||align="right"| P5 (pCMV_mVenus_YFP) |
| - | |-
| + | |
| - | | align="left" | DNA (500ng) ||align="left"| 1,4||align="left"|1,4||align="left"| 1,4||align="left"| 1,4
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"| 1||align="left"| 1||align="left"| 1||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1||align="left"| 1||align="left"| 1||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 1 NgoMIV ||align="left"|0,5||align="left"| 0,5||align="left"|0,5||align="left"|0,5
| + | |
| - | |-
| + | |
| - | | align="left" | H<sub>2</sub>O ||align="left"|6,1||align="left"| 6,1||align="left"| 6,1||align="left"| 6,1
| + | |
| | |- | | |- |
| - | | align="left" | '''Total volume''' ||align="left"|10||align="left"| 10||align="left"| 10||align="left"| 10 | + | | concentration (ng/µl)|| align="right" | 457||align="right"|470,8|| align="right" | 477,29 |
| | |} | | |} |
| - | <br />
| |
| - |
| |
| - | <br />
| |
| - | [[image:Freiburg10_Kopfkratzen.gif|thumb|right| Bitte schneid mich aus !]]
| |
| | | | |
| - | [[file:Freiburg10 test digestion NgoMIV.jpg]]
| + | ===14. Labortag 25.05.2010=== |
| | | | |
| - | According to the gel, NgoMIV SDM was successful in clones 1 and 2. Clone 3 well reveales 2 bands of unknown origin. Samples 1 and 3 were sent for sequencing.
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pCMV_mVenus_YFP + pAAV_MCS</b></p>==== |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Picking clones of pSB1C3_Affibody_linker and pSB1C3_ß-Globin_YFP_hGH_rITR </b></p>====
| + | <p><b>Investigators: Kira, Anna, Volker, Jessica</b></p> |
| - | <b>Investigator: Anna </b>
| + | |
| | | | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: A 1:1000 dilution of pSB1C3_Affibody_GSAT-Linker was prepared. <br/>
| + | <br> |
| - | <b>To do</b>: Mini-Prep of pSB1C3_Affibody_(Short/Middle/Long and SEG-Linker) and pSB1C3_ß-Globin_YFP_hGH_rITR. Picking clones of pSB1C3_Affibody_GSAT-Linker. </p>
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Test digestion of pSB1C3_SDM_SspI_Bla14FM (P222), pSB1C3_BLA (P286, P288) </b></p>====
| + | <li>plasmid: insert: pCMV_mVenus_YFP; number: P5 production date: 21.05.2010 origin: ____ |
| - | <b>Investigator: Achim</b> | + | <li>plasmid: vector: pAAV_MCS; number: P? production date: ____ origin: ____ |
| | + | <li>new vector name: pAAV_mVenus_YFP <br> |
| | | | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>New test digestion of two SDM-attempts, this time we also digested the original vector containing the PvuII restriction site to see differences.</p> | + | <br> |
| - | <br/>
| + | '''1st try: |
| | | | |
| | + | <li>buffer used:3 ; Restriction-enzymes used: Enzyme NotI (no. Lab:46) |
| | + | <li>DNA concentration (vector): 360 ng/µl ; DNA concentration (insert): 477 ng/µl |
| | | | |
| - | [[File:Freiburg10 Test digestion of pSB1C3 BLA final 24 08 2010.jpg|800px]]
| |
| - |
| |
| - |
| |
| - |
| |
| - | *The original vector is being cut as expected: the uncut vector can be seen in different conformations, the linearized vector shows one distinct band and the digestions cutting out BLA show a band at ~900 bp. Our SDM attempts show inconsistent results, not only does the Pvu site still seem to be in the backbone, there are no more proper insert bands visible either. We therefore conclude that either the DpnI digestion or the dilution of the transformation must have gone wrong (differing plasmids...). Tomorow we'll try a new cloning approach using an old vector with deleted Pvu restriction site and MscI & XbaI digestion. In case this also fails we also yet have to prep and test digest the repetition of the mutagenesis which was carried out by Stefan.
| |
| - |
| |
| - |
| |
| - | ===100. labday 25.08.2010===
| |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pSB1C3_Rep40, pSB1C3_Rep68 and pMA_RC_insert </b></p>====
| |
| - | '''Investigator: Chris L.'''<br>
| |
| - | *buffer used: 2; Restriction-enzymes used: Enzyme 1: HindIII ; Enzyme 2: BstEII
| |
| - | Plasmids:
| |
| - | <ul><li>pSB1C3_Rep40 '''P231'''</li>
| |
| - | <li>pSB1C3_Rep68 '''P266'''</li>
| |
| - | Insert:
| |
| - | <li>pMA_RC_insert '''P190'''</li>
| |
| - | </ul>
| |
| - | <br />
| |
| | {| border="1" | | {| border="1" |
| - | | align="left" | '''Components''' ||align="left"| '''P190/µL''' ||align="left"| '''P231/µL''' ||align="left"| '''P266/µL''' | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pAAV_MCS) / µl |
| | |- | | |- |
| - | | align="left" | DNA ||align="left"| 5.9 ||align="left"| 6.4 ||align="left"| 5
| + | | DNA || align="right" | 4.2||align="right"|3.5 |
| | |- | | |- |
| - | | align="left" | BSA (10x) ||align="left"| 2 ||align="left"|2||align="left"| 2
| + | | BSA (10x) || align="right" |3||align="right"|3 |
| | |- | | |- |
| - | | align="left" | Buffer 2 (10x) ||align="left"| 2 ||align="left"| 2||align="left"| 2
| + | | Buffer 3 (10x)|| align="right" |3||align="right"|3 |
| | |- | | |- |
| - | | align="left" | Enzyme 1 HindIII ||align="left"| 1 ||align="left"| 1 ||align="left"| 1
| + | |Enzyme: NotI (no.Lab:46)|| align="right" |1||align="right"|1 |
| | |- | | |- |
| - | | align="left" | Enzyme 2 BstEII ||align="left"| 1 ||align="left"| 1 ||align="left"| 1 | + | |H2O|| align="right" |15.3||align="right"|15.3 |
| | |- | | |- |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 8.1 ||align="left"| 7.6 ||align="left"| 9
| + | |'''Total volume'''|| align="right" |<b>26.4</b>||align="right"|<b>25.8</b> |
| - | |-
| + | |
| - | | align="left" | '''Total volume''' ||align="left"| 20 ||align="left"| 20||align="left"| 20
| + | |
| | |} | | |} |
| - | <br />
| + | |
| - | *Incubation: 90 minutes; 37°C with HindIII<br>
| + | <li> Incubation: 1 1/2 h at 37°C |
| - | *Incubation: 90 minutes; 60°C with BstEII<br>
| + | |
| - | <br />
| + | |
| - | '''Agarosegel'''
| + | |
| - | <br />
| + | |
| - | 0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED, 5 min at 90 Volt, 40 min at 115 Volt
| + | |
| - | <br />
| + | |
| - | [[File:Freiburg 10 pSB1C3 Rep40 pSB1C3 Rep68 pMA RC Insert.png|700px]]
| + | |
| - | <br />
| + | |
| - | '''Concentrations''' measured via NanoDrop:
| + | |
| - | *pSB1C3_Rep40: 81.97 ng/µl
| + | |
| - | *pSB1C3_Rep68: 48,68 ng/µl
| + | |
| - | *RC_Insert: 3,60 ng/µl
| + | |
| | <br> | | <br> |
| - | '''T4 Ligation of pSB1C3_Rep40 with RC_Insert'''<br> | + | '''note: too little water was added |
| - | Volume insert: 6,54 µl<br>
| + | |
| - | Volume vector: 1,46 µl<br>
| + | |
| | <br> | | <br> |
| - | '''T4 Ligation of pSB1C3_Rep68 with RC_Insert'''<br>
| |
| - | Volume insert: 5,46 µl<br>
| |
| - | Volume vector: 2,54 µl<br>
| |
| | <br> | | <br> |
| - | '''Trafo''' was prepared with XL1blue and Cm | + | 1% Agarose gel |
| | + | <br> |
| | + | <li>1% agarose gel was prepared, gel ran for 45 minutes(119 V) |
| | + | <br> |
| | + | <br> |
| | + | '''2nd try:''' |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Picking clones of pAAV_RC-ins_rep_cap and pCeruleanVP1up</b></p>====
| + | <li>buffer used:4 ; Restriction-enzymes used: Enzyme NotI (no. Lab:159) |
| - | <b>Investigator: Chris L. </b> | + | <li>DNA concentration (vector): 360 ng/µl ; DNA concentration (insert): 477 ng/µl |
| | | | |
| - | <b>To do</b>: Mini-Prep of pAAV_RC-ins_rep_cap and pCeruleanVP1up.
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cloning of pSB1C3_6xHis with middlelinker and pSB1C3_6xHis with pGA14_mVenus_YFP</b></p>====
| |
| - | '''Investigator: Jessica'''<br>
| |
| - | *buffer used: 4; Restriction-enzymes used: Enzyme 1: AgeI ; Enzyme 2: PstI ; Enzyme3: NgoMIV
| |
| - | <ul><li>Plasmids:</li>
| |
| - | <ul><li>pSB1C3_6xHis '''P84'''</li>
| |
| - | <li>pGA14_middle linker '''P65'''</li>
| |
| - | <li>GA14_mVenus_YFP '''P60'''</li>
| |
| - | </ul>
| |
| - | </ul>
| |
| - |
| |
| - |
| |
| - | <br />
| |
| | {| border="1" | | {| border="1" |
| - | | align="left" | '''Components''' ||align="left"| '''Mastermix''' ||align="left"| '''P84/µL''' ||align="left"| '''P65/µL''' ||align="left"| '''P60/µL''' | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pAAV_MCS) / µl |
| - | |-
| + | |
| - | | align="left" | DNA ||align="left"| -||align="left"| 13,91||align="left"| 13,52||align="left"| 8,93
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"|-||align="left"|2||align="left"| 2||align="left"| 2
| + | |
| | |- | | |- |
| - | | align="left" | Buffer 4 (10x) ||align="left"| -||align="left"| 2||align="left"| 2||align="left"| 2 | + | | DNA || align="right" | 4.2||align="right"|3.5 |
| | |- | | |- |
| - | | align="left" | Enzyme 1 AgeI ||align="left"|-||align="left"| 1||align="left"| -||align="left"| - | + | | BSA (10x) || align="right" |3||align="right"|3 |
| | |- | | |- |
| - | | align="left" | Enzyme 2 PstI ||align="left"| -||align="left"| 1||align="left"| 1 ||align="left"| 1 | + | | Buffer 4 (10x)|| align="right" |3||align="right"|3 |
| | |- | | |- |
| - | | align="left" | Enzyme 3 NgoMIV ||align="left"| -||align="left"| -||align="left"| 1||align="left"| 1
| + | |Enzyme: NotI (no.Lab:159)|| align="right" |1,5||align="right"|1,5 |
| | |- | | |- |
| - | | align="left" | H<sub>2</sub>O ||align="left"| -||align="left"| 0,09||align="left"| 0,48||align="left"| 5,07 | + | |H2O|| align="right" |18,3||align="right"|19 |
| | |- | | |- |
| - | | align="left" | '''Total volume''' ||align="left"| -||align="left"| 20||align="left"| 20||align="left"| 20
| + | |'''Total volume'''|| align="right" |<b>30</b>||align="right"|<b>30</b> |
| | |} | | |} |
| - | <br />
| |
| | | | |
| - | *Incubation:110 minutes; 37°C<br>
| + | <li> Incubation: 1 1/2 h at 37°C |
| - | '''Agarosegel'''
| + | |
| - | <br />
| + | |
| - | 0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED, at 120 Volt
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| | | | |
| - |
| |
| - | <br />
| |
| - | [[File:Freiburg10 gel cut n terminus1.jpg|500px|left|thumb|]]
| |
| | <br> | | <br> |
| | + | 1% Agarose gel |
| | <br> | | <br> |
| | + | <li>1% agarose gel was repared, gel ran for 45 minutes(119 V) |
| | <br> | | <br> |
| | + | |
| | + | <font color=#FF00FF>Results: unexpected sizes of fragments (see protocol), try again next day with an ethidium bromid gel</font> |
| | <br> | | <br> |
| | <br> | | <br> |
| | <br> | | <br> |
| - | <br> | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Production of chemical competent E.coli</b></p>==== |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | '''concentrations''' measured via NanoDrop:
| + | |
| - | *SB1C3_6xHis: 1,7 ng/µl
| + | |
| - | *middle linker: 3,8 ng/µl
| + | |
| - | *mVenus_YFP: 3,2 ng/µl
| + | |
| - | <br>
| + | |
| - | '''T4 Ligation pSB1C3_6xHis_middlelinker'''<br>
| + | |
| - | Volume insert: 0,25 µl<br>
| + | |
| - | Volume vector: 7,75 µl<br>
| + | |
| - | <br>
| + | |
| - | '''T4 Ligation pSB1C3_6xHis_mVenus_YFP'''<br>
| + | |
| - | Volume insert: 2,64 µl<br>
| + | |
| - | Volume vector: 5,36 µl<br>
| + | |
| - | <br>
| + | |
| - | '''Trafo''' was prepared with XL1blue and Cm
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of pSB1C3_6xHis</b></p>====
| + | <p><b>Investigators: Patrick and Jessica</b></p> |
| - | '''Investigator: Jessica'''<br>
| + | |
| - | '''P84''' was sequenced for continuation of working with it
| + | |
| | <br> | | <br> |
| | + | competent E.coli XL-1-blue and competent E.coli BL21 produced according to the standard-protocoll [[Media:production of competent E.coli.pdf]]<br> |
| | <br> | | <br> |
| | <br> | | <br> |
| | | | |
| - | [[File:Freiburg10 pSB1C3_6xHis.jpg|900px]]
| |
| - | <br>
| |
| - | * '''pSB1C3_6xHis P84 is ready to use, but is empty. a new tube will be made tomorrow'''
| |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Inoculation of pGA14_middlelinker and pSB1C3_6xHis clone1 </b></p>==== | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Design of MCS-Oligos</b></p>==== |
| - | '''Investigator: Jessica'''<br>
| + | <p><b>Investigators: Bea, Adrian, Hanna, Sven</b></p> |
| - | *both tubes ('''P65''' and '''P84''')are empty --> inoculation with glycerolstocks
| + | |
| - | **'''B48''' pGA14_middlelinker was inoculated with 10ml DYT and 10µl Amp
| + | |
| - | **'''B64''' pSB1C3_6xHis clone1 was inoculated with 10ml DYT and 10µl Cm
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;">'''Cloning of pSB1C3_SspI_BLA and pSB1C3_CFP_PvuII'''</p>====
| + | |
| - | | + | |
| - | <b> Investigator: Achim</b> | + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>:I ligated a sequence from pSB1C3_CFP_PvuII missing the PvuII restriction site into pSB1C3_SspI_BLA to obtain a pSB1C3 vector without PvuII in the cat marker.</p>
| + | |
| - | | + | |
| - | Vector: <br>
| + | |
| - | *pSB1C3_SspI_BLA (p222): c= 308,32 ng/µl<br>
| + | |
| - | Insert: <br>
| + | |
| - | *pSB1C3_CFP_PvuII (p129): c= 264,86 ng/µl<br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | components || align="right" |Vector || align="right" |Insert
| + | |
| - | |-
| + | |
| - | | DNA || align="right" |3,8 || align="right" | 4,9
| + | |
| - | |-
| + | |
| - | | BSA (10x) || align="right" |2|| align="right" | 2
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x)|| align="right" |2 || align="right" | 2
| + | |
| - | |-
| + | |
| - | |Enzyme MscI || align="right" |1 || align="right" | 1
| + | |
| - | |-
| + | |
| - | |Enzyme XbaI || align="right" |1|| align="right" | 1
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |10,2 || align="right" | 9,1
| + | |
| - | |-
| + | |
| - | |'''Total volume '''|| align="right" |20 || align="right" | 20
| + | |
| - | |}
| + | |
| | <br> | | <br> |
| | + | In order to replace the multiple cloning site of the pAAV_MCS vector from Stratagene by RFC25, two oligos were designed. After their hybridization the overhangs correspond to ClaI- and BglII restriction sites. A digestion with BglII and ClaI will be performed with pAAV-MCS and then will be ligated with the designed oligos.<br> |
| | + | The oligos (see link) were ordered at Sigma-Aldrich. Estimated shipment: 31.05.2010 .<br> |
| | | | |
| - | *Gelextraction:
| |
| - | 0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 110 Volt, running time:45 <br />
| |
| - | Marker: GeneRuler ladder mix (Fermentas)
| |
| - | <br />
| |
| - | <br />
| |
| - | <br />
| |
| - | <br />
| |
| | | | |
| - | [[Image:Freiburg10 25082010achim.JPG|400px]] | + | [[File:Freiburg10 Oligos MCS RFC25 for pAAV.pdf]] |
| - | <br />
| + | |
| | | | |
| | + | ===15. Labortag 26.05.2010=== |
| | | | |
| | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Oligos (PstI + MCS)</b></p>==== |
| | | | |
| - | *c(Insert)= 15,66 ng/µl; size: 730 bp<br />
| + | <p><b>Investigators: Bea, Hanna, Jessica</b></p> |
| - | *c(Vector)= 14,42 ng/µl; size: 2200 bp
| + | |
| | | | |
| - | <br />
| |
| - |
| |
| - | *Ligation of PCR products and vector:
| |
| - |
| |
| - | For the Ligation 1µl T4 buffer (2x) and 1µl T4 ligase were used. Incubation time: 60 min due to blunt end ligation
| |
| - | <br />
| |
| - | {| border="1"
| |
| - | | ''' ''' || align="right" |'''vector /µl''' || align="right" |'''insert /µl'''
| |
| - | |-
| |
| - | | pSB1C3_BLA || align="right" |0,86 || align="right" |7,14
| |
| - | |-
| |
| - | |}
| |
| - | <br />
| |
| - |
| |
| - | *Transformation:
| |
| - |
| |
| - | The transformation was done following the standard protocol using XL1 blue cells.<br />
| |
| - | <br />
| |
| - |
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Two clones were picked, but because we already obtained our plasmid via SDM in the meantime, no prep was performed. Glycerol stocks of the two clones were created just in case, B264 and B265.</p>
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Preps and test digestion of pSB1C3_Affibody_Middle-Linker, pSB1C3_Affibody_Short-Linker, pSB1C3_Affibody_SEG-Linker, pSB1C3_Affibody_Long-Linker and pSB1C3_betaglobin_mVenus_hGH_rITR</b></p>====
| |
| - | <b>Investigator: Stefan</b><br>
| |
| | <ul> | | <ul> |
| - | <b>Glycerol stocks were prepared:</b>
| + | <li>Repeat cloning of pAAV-MCS + pCMV_mVenus_YFP --> <b>Goal: pAAV_mVenus_YFP</b><br> |
| - | <li>B240 = pSB1C3_Affibody_Middle-Linker clone1</li> | + | |
| - | <li>B241 = pSB1C3_Affibody_Middle-Linker clone2</li> | + | |
| - | <br /> | + | |
| | | | |
| - | <li>B242 = pSB1C3_Affibody_Short-Linker clone 1</li>
| + | Cloning did not work (see lab day 25.05.2010) either with NotI or with NotI-HF. Gel did not show any proper band at the expected size. <br> |
| - | <li>B243 = pSB1C3_Affibody_Short-Linker clone 2</li>
| + | There will be used EtBr instead of gelred in the agarose gel and the incubation time of digestion will be prolonged. Further details are followed. <br> |
| - | <br />
| + | NOTE: There are three restriction sites of NotI in the pCMV-mVenus_YFP. If cloning with NotI, the polyA hGH signal will be deleted. therefore cloning with NotI is '''not''' possible . |
| - | | + | |
| - | <li>B246 = pSB1C3_Affibody_SEG-Linker clone1</li>
| + | |
| - | <li>B247 = pSB1C3_Affibody_SEG-Linker clone2</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>B248 = pSB1C3_Affibody_Long-Linker clone 1</li>
| + | |
| - | <li>B249 = pSB1C3_Affibody_Long-Linker clone 2</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>B244 = pSB1C3_betaglobin_mVenus_hGH_rITR clone 1</li>
| + | |
| - | <li>B245 = pSB1C3_betaglobin_mVenus_hGH_rITR clone 2</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | | + | |
| - | <b>Mini-Prep following the standard protocol</b>
| + | |
| - | <li>P290 = pSB1C3_Affibody_Middle-Linker clone1 c= 227,36 ng/µl</li>
| + | |
| - | <li>P291 = pSB1C3_Affibody_Middle-Linker clone2 c= 219,05 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>P292 = pSB1C3_Affibody_Short-Linker clone 1 c= 196,63 ng/µl</li>
| + | |
| - | <li>P293 = pSB1C3_Affibody_Short-Linker clone 2 c= 224,76 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>P296 = pSB1C3_Affibody_SEG-Linker clone1 c= 218,68 ng/µl</li>
| + | |
| - | <li>P297 = pSB1C3_Affibody_SEG-Linker clone2 c= 229,66 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>P298 = pSB1C3_Affibody_Long-Linker clone 1 c= 229,73 ng/µl</li>
| + | |
| - | <li>P299 = pSB1C3_Affibody_Long-Linker clone 2 c= 202,05 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <li>P294 = pSB1C3_betaglobin_mVenus_hGH_rITR clone 1 c= 425,14 ng/µl</li>
| + | |
| - | <li>P295 = pSB1C3_betaglobin_mVenus_hGH_rITR clone 2 c= 328,28 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | </ul>
| + | |
| - | <b>Test digestion</b><br>
| + | |
| - | <ul>
| + | |
| - | <li>Restriction-enzymes used: for P290-P293 and P296-P299: NotI ; for P294-P295: NgoMIV and PstI </li>
| + | |
| - | </ul><br />
| + | |
| - | <b>'''comment:'''</b> The same amount of ingredients were used for P290-P293 and P296-P299, therefore these approaches will be merged into the chart. The same goes for P294-P295.
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| '''P290-P293 and P296-P299''' ||align="left"| '''P294-P295'''
| + | |
| - | |-
| + | |
| - | | align="left" | DNA ||align="left"| 1 ||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"| 1 ||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1 ||align="left"| 1
| + | |
| - | |-
| + | |
| - | | align="left" | NotI ||align="left"| 0,5 ||align="left"| -
| + | |
| - | |-
| + | |
| - | | align="left" | NgoMIV ||align="left"| - ||align="left"| 0,5
| + | |
| - | |-
| + | |
| - | | align="left" | PstI ||align="left"| - ||align="left"| 0,5
| + | |
| - | |-
| + | |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 6,5 ||align="left"| 6
| + | |
| - | |-
| + | |
| - | | align="left" | <b>Total volume</b> ||align="left"| <b>10</b> ||align="left"| <b>10</b>
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | Incubation time: 70 minutes; Incubation temperature: 37°
| + | |
| - | <br />
| + | |
| - | 0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , running time:5 minutes at 90 Volt and 50 minutes at 115 Volt <br />
| + | |
| - | 2µl loading dye (6x) for the sample, Marker: GeneRuler ladder mix (Fermentas)
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | [[File:Freiburg10 test digestion affi+linker right assembly.jpg|500px]]
| + | |
| - | <br /> | + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Sizes of fragments look like expected. Clone 1 of each plasmid will be sent for sequencing tomorrow.</p>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequenzing evaluation of SDM NgoMIV</b></p>====
| + | |
| - | <b>Investigator: Kira</b><br>
| + | |
| - |
| + | |
| - | 2 samples have been sent for sequencing yesterday. According to the data, both samples do not contain any NgoMIV restriction site anymore. <br />
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | [[File:Freiburg10 sample 1, seq 1.jpg|500px]]
| + | |
| - | | + | |
| - | | + | |
| - | [[File:Freiburg10 sample 3, seq2.jpg|500px]]
| + | |
| - | | + | |
| - | ===101. labday 26.08.2010===
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Preps of pGA14_middle linker and pSB1C3_6xHis clone 1</b></p>====
| + | |
| - | <b>Investigator: Jessica</b><br>
| + | |
| - | <ul>
| + | |
| - | Glycerol stocks were not prepared because I have inoculated from glycerol stocks
| + | |
| | <br> | | <br> |
| - |
| |
| - | <b>Mini-Prep following the standard protocol</b>
| |
| - | <li>P301 = pGA14_middle linker (= '''P65''')<br>
| |
| - | c=305,7</li>
| |
| - | <br />
| |
| - | <li>P300 = pSB1C3_6xHis clone 1 (= '''P84''')<br>
| |
| - | c=177,5</li>
| |
| - |
| |
| - | <br />
| |
| - | </ul>
| |
| - |
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Preps and test digestion of pAAV_RC-ins_rep_cap, pCeruleanVP1up and pAAV_RC_ins-rep-cap.</b></p>====
| |
| - |
| |
| - |
| |
| - |
| |
| - | <b>Investigator: Chris L.</b><br>
| |
| - | <ul>
| |
| - | <b>Glycerol stocks were prepared:</b>
| |
| - | <li>B250 = pCerulean_VP1up clone1</li>
| |
| - | <li>B251 = pCerulean_VP1up clone2</li>
| |
| - | <li>B252 = pCerulean_VP1up clone3</li>
| |
| - | <li>B253 = pCerulean_VP1up clone4</li>
| |
| - | <li>B254 = pSB1C3_VCK_Bla clone1</li>
| |
| - | <li>B255 = pSB1C3_VCK_Bla clone2</li>
| |
| - | <li>B256 = pSB1C3_VCK_Bla clone3</li>
| |
| - | <li>B257 = pSB1C3_VCK_Bla clone4</li>
| |
| - | <li>B262 = pAAV_RC_ins-rep-cap clone1</li>
| |
| - | <li>B263 = pAAV_RC_ins-rep-cap clone2</li>
| |
| - | <br />
| |
| - | <b>Mini-Prep following the standard protocol</b>
| |
| - | <li>P302 = pCerulean_VP1up clone1<br>c=410,70 ng/µl</li>
| |
| - | <li>P303 = pCerulean_VP1up clone2<br>c=387,78 ng/µl</li>
| |
| - | <li>P304 = pCerulean_VP1up clone3<br>c=321,04 ng/µl</li>
| |
| - | <li>P305 = pCerulean_VP1up clone4<br>c=350,34 ng/µl</li>
| |
| - | <li>P306 = pSB1C3_VCK_Bla clone1<br>c=82,53 ng/µl</li>
| |
| - | <li>P307 = pSB1C3_VCK_Bla clone2<br>c=92,70 ng/µl</li>
| |
| - | <li>P308 = pSB1C3_VCK_Bla clone3<br>c=82,22 ng/µl</li>
| |
| - | <li>P309 = pSB1C3_VCK_Bla clone4<br>c=105,34 ng/µl</li>
| |
| - | <li>P314 = pAAV_RC_ins-rep-cap clone1<br>c=517,04 ng/µl</li>
| |
| - | <li>P315 = pAAV_RC_ins-rep-cap clone2<br>c=444,33 ng/µl</li>
| |
| - | <br />
| |
| - | </ul>
| |
| - | <b>Test digestion:</b>
| |
| | <br> | | <br> |
| - | {| border="1"
| + | <li>Test digestion of pCMV-mVenusYFP with PstI and MluI <br> |
| - | | components || align="right" |volume of '''P302'''/µl ||volume of '''P303'''/µl||volume of '''P304/'''µl ||volume of '''P305'''/µl ||volume of '''P306'''/µl||volume of '''P306'''/µl||volume of '''P306'''µl ||volume of '''P307'''/µl||volume of '''P307'''µl ||volume of '''P307'''/µl||volume of '''P308'''/µl||volume of '''P308'''/µl ||volume of '''P308'''/µl ||volume of '''P309'''/µl ||volume of '''P309'''/µl ||volume of '''P309'''/µl||volume of '''P314'''/µl ||volume of '''P315'''/µl
| + | |
| - | |-
| + | |
| - | | DNA || 1 || 1 || 1 || 1 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 2 || 1 || 1
| + | |
| - | |-
| + | |
| - | | BSA (10x) ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x) ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||1 ||- ||-
| + | |
| - | |-
| + | |
| - | | Buffer 2 (10x) ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-||- ||1 ||1
| + | |
| - | |-
| + | |
| - | |Enzyme NotI ||- ||- ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||- ||-
| + | |
| - | |-
| + | |
| - | |Enzyme SspI ||- ||- ||- ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||0,5 ||- ||- ||-
| + | |
| - | |-
| + | |
| - | |Enzyme SalI ||- ||- ||- ||- ||- ||- ||0,5 ||- ||- ||0,5 ||-||- ||0,5 ||- ||- ||0,5 ||- ||-
| + | |
| - | |-
| + | |
| - | |Enzyme BamHI ||- ||- ||- ||- ||- ||- ||0,5 ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-||-
| + | |
| - | |-
| + | |
| - | |Enzyme PvuII ||- ||- ||- ||- ||- ||- ||0,5 ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-||-
| + | |
| - | |-
| + | |
| - | |Enzyme Acc65I ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-||- ||0,75 ||0,75
| + | |
| - | |-
| + | |
| - | |Enzyme XcmI ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-||- ||0,5 ||0,5
| + | |
| - | |-
| + | |
| - | |Enzyme PstI ||0,5 ||0,5 ||0,5 ||0,5 ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-
| + | |
| - | |-
| + | |
| - | |Enzyme EcoRI ||0,5 ||0,5 ||0,5 ||0,5 ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||- ||-
| + | |
| - | |-
| + | |
| - | |H2O ||6 ||6 ||6 ||6 ||5,5 ||5 ||5 ||5,5 ||5 ||5 ||5,5 ||5||5 ||5,5 ||5 ||5 ||5,75 ||5,75
| + | |
| - | |-
| + | |
| - | |'''Total volume /µl'''||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10 ||10
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | Incubation time: 1 h, Incubation temperature: 37°
| + | |
| - | <br />
| + | |
| - | Preparation of gel:<br />
| + | |
| - | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 50 minutes
| + | |
| - | <br /> | + | |
| | | | |
| - | [[File:Freiburg10 test digestion pSB1C3 VCK Bla+pCerulean Vp1up.jpg]]
| + | <b>Digestion</b> |
| - | | + | |
| - | <b>Results:</b> <p style="color:#00bbff;">pSB1C3_VCK_Bla: Test digestion looks like expected. Clone 4 was sent for sequencing. <br />
| + | |
| - | pCerulean_VP1up: Gel looks well as well. clone 2 was sent for sequencing. <br/>
| + | |
| - | pAAV_RC_ins-rep-cap: Test digestion looks strange. Just one big band with more than 10000 bp and one very small with 150 bp. Maybe the restriction enzymes didn´t cut. (@Christian: please insert picture)</p>
| + | |
| - | | + | |
| - | ===102. labday 27.08.2010===
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of pSB1C3_Bla_final (P309)and pCerulean_VP1up (P303)</b></p>====
| + | |
| - | <b>Investigator: Bea </b>
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#cc0033;"><I><b>GENERAL COMMENT:</b> pSB1C3_Bba_final (P309, SB_9) and pCerulean_Vp1up (P303; SB_10) were sent for sequencing. In the case of SB1C3_Bba_final, which means that the whole vector was assembled from the buttom,the purpose was to test the designed primers. In the other case we wanted to verify the insertion of Vp1up inot pCerulean. </i> </p>
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pSB1C3_leftITR_CMV_betaglobin_mVenus_hGH_rightITR.</p>
| + | |
| - | <ul>
| + | |
| - | <li>Primer used: </li>
| + | |
| - | <ul>
| + | |
| - | <li>pSB1C3_Bba_seq_for</li>
| + | |
| - | <li>pSB1C3_Bba_seq_rev</li>
| + | |
| - | </ul>
| + | |
| - | <li>Plasmid sequenced: P?? </li>
| + | |
| - | <li>Sequence sample: ??</li>
| + | |
| - | <li>Stored in Geneious-folder: BioBricks --> final parts</li>
| + | |
| - | </ul>
| + | |
| - | | + | |
| - | <gallery widths=600px heights=400px perrow=2 caption="pSB1C3_leftITR_CMV_betaglobin_mVenus_hGH_rightITR">
| + | |
| - | Image:Freiburg10 Seqanalysis pSB1C3 Bba final 2010 27 08.jpg
| + | |
| - | </gallery>
| + | |
| - | <br />
| + | |
| - | <b>Results:</b> <p style="color:#00bbff;">Results look good. The forward primer worked, still waiting for the results of the reverse primer. The scar inbetween the CMV promoter and the beat globin intron corresponds to the expected result.</p>
| + | |
| - | | + | |
| - | <br />
| + | |
| - | <b>Next steps: Wwait until sequencing results of the reverse primer can be confirmed aswell. If that will not be the case, order new sequensing primer.</b>
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | | + | |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pCerulean_Vp1up.</p>
| + | |
| - | <ul>
| + | |
| - | <li>Primer used: CMV-F</li>
| + | |
| - | <li>Plasmid sequenced: P303 </li>
| + | |
| - | <li>Sequence sample: ??</li>
| + | |
| - | <li>Stored in Geneious-folder: N-terminal Targeting --> pCerulean_Vp1up</li>
| + | |
| - | </ul>
| + | |
| - | | + | |
| - | <gallery widths=600px heights=400px perrow=2 caption="pCerulean_Vp1up">
| + | |
| - | Image:Freiburg10 Seqanalysis pCerulean Vp1up 2010 27 08.jpg
| + | |
| - | </gallery>
| + | |
| - | <br />
| + | |
| - | <b>Results:</b> <p style="color:#00bbff;">Results are good. Insertion of the Vp1up region can be confirmed.</p>
| + | |
| - | | + | |
| - | <br />
| + | |
| - | <b>Next steps: Fuse pCerulean_Vp1up to the NLS. </b>
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | | + | |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Hybridisation and cloning of loop insertions</b></p>====
| + | |
| - | <b>Investigator: Achim, Volker</b>
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px;"><b>We hybridized the oligos for the different loop insertions and cloned them into the pSB1C3_BLA backbone.</b> </p>
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#cc0033;"><b>Hybridisation</b> </p>
| + | |
| - | *4 Inserts contained overlapping ends and had to be filled up using Klenov fragments. We therefore added Klenow buffer and dNTPs to those samples.
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| '''453 BAP''' ||align="left"| '''587 BAP''' ||align="left"| '''587 KO BAP'''||align="left"| '''453 RGD'''||align="left"| '''587 RGD'''||align="left"| '''587 KO RGD'''||align="left"| '''453 HIS'''||align="left"| '''587 HIS'''||align="left"| '''587 KO HIS'''||align="left"| '''587 KO EMPTY'''||align="left"| '''453 Z34C'''||align="left"| '''587 Z34C'''||align="left"| '''587 KO Z34C'''||align="left"| '''587 KO Z34C SPACER'''
| + | |
| - | |-
| + | |
| - | | align="left" | Oligo 1||align="left"|O124: 10 ||align="left"|O126: 10 ||align="left"|O128: 10 ||align="left"|O130: 10 ||align="left"|O132: 10 ||align="left"|O134: 10 ||align="left"|O135: 10 ||align="left"|O137: 10 ||align="left"|O139: 10 ||align="left"|O141: 10 ||align="left"|O143: 10 ||align="left"|O145: 10 ||align="left"|O147: 10 ||align="left"|O149: 10
| + | |
| - | |-
| + | |
| - | | align="left" | Oligo 2 ||align="left"|O125: 10 ||align="left"|O127: 10 ||align="left"|O129: 10 ||align="left"|O131: 10 ||align="left"|O133: 10 ||align="left"|O151: 10 ||align="left"|O136: 10 ||align="left"|O138: 10 ||align="left"|O140: 10 ||align="left"|O142: 10 ||align="left"|O144: 10 ||align="left"|O146: 10 ||align="left"|O148: 10 ||align="left"|O150: 10
| + | |
| - | |-
| + | |
| - | | align="left" | TrisHCl pH8||align="left"|4||align="left"|4||align="left"|4||align="left"|4||align="left"|4||align="left"|4 ||align="left"|4||align="left"|4||align="left"|4||align="left"|4||align="left"|-||align="left"|-||align="left"|-||align="left"|-
| + | |
| - | |-
| + | |
| - | | align="left" | 5mM MgCl2||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|8 ||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|-||align="left"|-||align="left"|-||align="left"|-
| + | |
| - | |-
| + | |
| - | | align="left" | Klenow Buffer||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|- ||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|4||align="left"|4||align="left"|4||align="left"|4
| + | |
| - | |-
| + | |
| - | | align="left" | dNTP Mix||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|- ||align="left"|-||align="left"|-||align="left"|-||align="left"|-||align="left"|1||align="left"|1||align="left"|1||align="left"|1
| + | |
| - | |-
| + | |
| - | | align="left" | H2O||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|8 ||align="left"|8||align="left"|8||align="left"|8||align="left"|8||align="left"|15||align="left"|15||align="left"|15||align="left"|15
| + | |
| - | |-
| + | |
| - | | align="left" | <b>Total volume</b>||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b> ||align="left"| <b>40</b>
| + | |
| - | |}
| + | |
| - | | + | |
| - | *Hybridisation was carried out according to standard protocoll
| + | |
| - | *Klenov fill-in reaction:
| + | |
| - | **Added 1 µl of NEB Klenow fragment to samples 11,12,13,14
| + | |
| - | **incubated for 1h @ 37°C
| + | |
| - | <p style="font-size:13px; color:#cc0033;"><b>Digestion of pSB1C3_BLA vector and Samples 11-14 </b> </p>
| + | |
| - | *We digested the standard vector with the 453 (Ssp/Sal) and the 587 (Bam/Pvu) standard. The samples that were filled in were also digested to create sticky ends.
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| '''V453''' ||align="left"| '''V587''' ||align="left"| '''11''' ||align="left"| '''12''' ||align="left"| '''13''' ||align="left"| '''14'''
| + | |
| - | |-
| + | |
| - | | align="left" | DNA ||align="left"| 2,5 ||align="left"| 2,5 ||align="left"| 3,5||align="left"| 3,5||align="left"| 3,5||align="left"| 3,5
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"| - ||align="left"| - ||align="left"| - ||align="left"| - ||align="left"| - ||align="left"| -
| + | |
| - | |-
| + | |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 2 ||align="left"| 2 ||align="left"| 2 ||align="left"| 2 ||align="left"| 2 ||align="left"| 2
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 1||align="left"| Ssp: 1 ||align="left"| Bam: 1 ||align="left"| Ssp: 1 ||align="left"| Bam: 1 ||align="left"| Bam: 1 ||align="left"| Bam: 1
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 2||align="left"| Sal:1 ||align="left"| Pvu: 1 ||align="left"| Sal: 1 ||align="left"| Pvu: 1 ||align="left"| Pvu: 1 ||align="left"| Pvu: 1
| + | |
| - | |-
| + | |
| - | | align="left" | H2O||align="left"| 13,5 ||align="left"| 13,5 ||align="left"| 12,5 ||align="left"| 12,5 ||align="left"| 12,5 ||align="left"| 12,5
| + | |
| - | |-
| + | |
| - | | align="left" | <b>Total volume</b> ||align="left"| <b>20</b> ||align="left"| <b>20</b> ||align="left"| <b>20</b>||align="left"| <b>20</b>||align="left"| <b>20</b>||align="left"| <b>20</b>
| + | |
| - | |}
| + | |
| - | <p style="font-size:13px; color:#cc0033;"><b>Gel Extraction </b> </p>
| + | |
| - | | + | |
| - | [[Image:Freiburg10 27082010achim.JPG|500px]]
| + | |
| - | <p style="font-size:13px; color:#cc0033;"><b>Ligation</b> </p>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning of pCerulean_VP1up_NLS</b></p>====
| + | |
| - | <b>Investigator: Anissa </b>
| + | |
| - | <p style="font-size:13px; color:red;"><b>Comment</b>:Cloning did not work, because no NLS-band could be seen in the gel... Will be repeated on monday in two new approaches. One time VP1up will be cloned into pSB1C3_NLS and recloned as a fusion-product into pCerulean. Another time the oligos of NLS will be hybridisized, cut with rescriction enzymes and purificated with the Qiaex II kit.Then the NLS will be ligated into the pCerulean_VP1up </p>
| + | |
| - | <br/>
| + | |
| - | *Digestion:
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | components || align="right" |Vector || align="right" |Insert
| + | |
| - | |-
| + | |
| - | | DNA || align="right" |2,6 || align="right" | 32
| + | |
| - | |-
| + | |
| - | | BSA (10x) || align="right" |1,5|| align="right" | -
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x)|| align="right" |1,5 || align="right" | 4
| + | |
| - | |-
| + | |
| - | |Enzyme || align="right" |1 PstI || align="right" | 2 PstI
| + | |
| - | |-
| + | |
| - | |Enzyme || align="right" |1 AgeI|| align="right" | 2 NgoMIV
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |7,4 || align="right" | -
| + | |
| - | |-
| + | |
| - | |'''Total volume '''|| align="right" |15|| align="right" | 40
| + | |
| - | |}
| + | |
| | <br> | | <br> |
| - | * Gel: that's only the picture of the 2% agarose gel for seperating the NLS. But no band could be seen.
| |
| - | [[image:Freiburg10 NLS.jpg|400px ]]
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>DARPin E_01</b></p>====
| |
| - | <b>Investigator: Bea </b>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#66bbaa;"><b>General overview of DARPins:</b></p>
| |
| - | Natural protein ankyrin repeat molecules are motifs which can be found in proteins. These motifs are mediating protein-protein interactions. This suggests that ankyrin repeat (AR) proteins can be used for designing binding molecules. In Kohl <i>et al.</i> (2003) and Binz et al. (2003) the authors designed a structural framework with fixed consensus regions and randomized positions of interacting residues.
| |
| - | <br />
| |
| - |
| |
| - | The repetitive nature of the ankyrin proteins allows modifications in their variable and modular binding surface. Therefore consensus sequences of natural ankyrin proteins were used to design novel and stable scaffolds for binding proteins.
| |
| - | Designed Ankyrin Proteins (DARPins) are well expressed, monomeric in solution, thermodynamically stable and have the ability to fold fast. In the publication of Steiner <i>et al.</i> (2008) screening libraries were created by useing the signal recognition particle (SRP) translocation pathway for phage display. The proteins containing the appropriate translocation signal sequence are efficiently displayed on filamentous phage particles. Screening for DARPins binding to specific target proteins like EGF-R (Erb1) were performed by phage ELISAs. The extracellular domain I-III of the receptor ErbB1 was fused to the Fc part of human IgG1 and used for selection. The previously described combinatorial N3C library was used as a template for SRP selection. After phage ELISA ans sequencing have been performed only one clone (E_01) could be selected with high-affinity binding characteristics. For broaden the diversity of ErbB1 DARPins three more clones were selected by epitope masking (E_67, E_68 and E_69). Binding and epitope localization experiments showed that all selected clones recognize a competing epitope as the dominant binder E_01, except of E_69 which recognizes an epitope not competing with E_01. The low diversity obtained for DARPins against ErbB1 suggests that the binding interfaces are well suited for dominant selection. Additionally the selected clones showed no cross-reactivity with other ErbB-family receptors (Figure 2C).
| |
| - | <br/>
| |
| - | <br/>
| |
| - | <gallery widths=400px heights=400px>
| |
| - | Image: Freiburg10 DARPin E 01 table Steiner et al.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | The dominant DARPpin E_01 has very high affinities to the target protein ErbB1 (Table 3) and can be used as a potential targeting molecule four our approach in fusing the DARPin to the N-terminal VP proteins.
| |
| - | <br />
| |
| - | <br/>
| |
| - | <p style="font-size:13px; color:#66bbaa;"><b>Overview of DARPinE_01 used in our approach: </b></p>
| |
| - | <gallery widths=600px heights=200px>
| |
| - | Image:Freiburg10 DARPin E 01.jpg
| |
| - | </gallery>
| |
| - | The designed ankyrin repeat protein used as a potential targeting molecule consists of three internal capping repeats and the C-and N-terminal capping repeats. Each internal repeat module comprises of one beta-turn and two hydrophobic alpha-helices. The potential interaction residues are located in the beta-turn the first alpha-helices of the AR-proteins. The complete nucleotide and corresponding amino acid sequence can be found in the appendix.
| |
| - | <gallery widths=400px heights=400px perrow=2>
| |
| - | Image: Freiburg10 DARPin E 01 structure.jpg
| |
| - | Image: Freiburg10 DARPin E 01 structure 2.jpg
| |
| - | </gallery>
| |
| - | <br/>
| |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Update VP1 insertion</b></p>====
| |
| - | <b>Investigator: Anissa, Bea </b>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#cc0033;"><b>Done:</b> </p>
| |
| | <ul> | | <ul> |
| - | <li> <b>pSB1C3_VP1up</b> was succesfully created by Achim. For details see labday from 2010-22-08.</li> | + | <li>experiment date: 26.05.2010 </li> |
| - | <li> <b>pCerulean_VP1up</b> was suceesfully created by Anissa. pCerulean was sequenced and the VP1up which contains the upstream 137 aa´s of the VP1 protein could be successfully cloned into the plasmid backbone pCerulean (obtained by performeing a PCR).</li> | + | <li>plasmid: pCMV_mVenus_YFP; number: P5; production date: 21.05.2010/ pCMV-MSC; number: P2; production date: </li> |
| - | <li><b>pSB1C3_NLS</b>Hybridization of the nuclear localization sequence (NLS) was successfully incorporated into the pSB1C3_CFP. </li> | + | <li>buffer used: 3/4; Restriction-enzymes used: Enzyme PstI (no. Lab:48) and Enyzme MluI (no. Lab:40) / NotI HF (no. Lab:159) </li> |
| - | <li>Today, Anissa tried to fuse the NLS to the VP1up sequence. Cutting the pSB1C3_NLS with NgomIV and PstI leads to a 45 nt fragment which normally can be resoluted in a 2% agarose gel. But no fragment could be detected in the gel (for further details see: Cloning of <b>pCerulean_VP1up_NLS</b>). | + | <li>DNA concentration plasmid: P5 477,3 ng/µl / P2 550ng/µl </li> |
| - | <br />
| + | |
| - | On monday another approach will be performed: The NLS oligos will be hybridized and digested with NgomIV and SpeI. With the Qiaex II Kit (protocol for desalting and concentrating DNA solutions),the hybrifized oligos will be purified. Parallel, another digestion approach will be conducted. </li>
| + | |
| | | | |
| - | <gallery widths=300px heights=400px perrow=2 caption="Overview">
| |
| - | Image:Freiburg10 Overview VP1 part1.jpg
| |
| - | Image:Freiburg10 Overview VP1 part2.jpg
| |
| - | </gallery>
| |
| - | </ul>
| |
| - | <p style="color:#00bbff;"> Next steps: </p>
| |
| - | <ul>
| |
| - | <li> We obtain pSB1C3_VP23_HSPG_ko by conducting a PCR with the pAAV_RC_final (which contains the integrated "cap". </li>
| |
| - | <li> Fuse the obtained construct pSB1C3_Targeting Molcule to the pSB1C3_VP23_HSPG_ko. </li>
| |
| - | <ul>
| |
| - | <li>Affibody Z<sub>EGFR:1907</sub></li>
| |
| - | <li>DARPin E_01</li>
| |
| - | <li>6xHis Tag</li>
| |
| - | <li>CFP</li>
| |
| - | </ul>
| |
| - | <li> Fuse pSB1C3_VP23_HSPG_ko_Targeting molecule to the construct pCerulean_VP1up_NLS.</li>
| |
| - | <li> Finally, for providing the possibilty to express obtained construct in trans: perform a site-directed mutagenesis with the pAAV_RC_final to remove the startcodon of VP1.</li>
| |
| - | </ul>
| |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of pSB1C3_BLA_final (P309)</b></p>====
| |
| - | <b>Investigator: Bea </b>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#cc0033;"><I><b>GENERAL COMMENT:</b> pSB1C3_BLA_final (P309) was sent for sequencing. the beta lactamse was inserted and two site-directed mutagenesis have been performed in order to delete the loop insertion restriction enzymes in the CAT marker.</i> </p>
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: Sequence analysis of pSB1C3_BLA_final containing the two deleted restriction sites (SspI and PvuII)in the CAT marker.</p>
| |
| - | <ul>
| |
| - | <li>Primer used: </li>
| |
| - | <ul>
| |
| - | <li>pQE-RP</li>
| |
| - | <li>VF-2</li>
| |
| - | </ul>
| |
| - | <li>Plasmid sequenced: P309 </li>
| |
| - | <li>Sequence sample: SB_9</li>
| |
| - | <li>Stored in Geneious-folder: pSB1C3_BLA</li>
| |
| - | </ul>
| |
| - |
| |
| - | <gallery widths=500px heights=400px perrow=2 caption="pSB1C3_BLA_final">
| |
| - | Image:Freiburg10 Seqanalysis pSB1C3 BLA final P309 SspI.jpg
| |
| - | Image:Freiburg10 Seqanalysis pSB1C3 BLA final P309 PvuII.jpg
| |
| - | Image:Freiburg10 Seqanalysis pSB1C3 BLA final P309.jpg
| |
| - | </gallery>
| |
| - | <br />
| |
| - | <b>Results:</b> <p style="color:#00bbff;">The two performed site-directed mutagenesis performed at the backbone of the pSB1C3 containing the beta-lactamase can be confrimed as it can be seen in the two pictures above. Additionally the BLA sequence was sequenced aswell as the insertion of the BLA which can be confirmed with no mutations.</p>
| |
| - |
| |
| - | <br />
| |
| - | <b>Next steps: This new plasmid can be used for further experiments. Clone the PCR prdouct of the pAAV_RC into the pSB1C3 and subclone the loop insertion motifs into the vector.</b>
| |
| - | <br />
| |
| - | <br />
| |
| - |
| |
| - | [http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/Laborjournal top of page]<br />
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Preps and test digestion of pSB1C3_6xHis_middlelinker and pSB1C3_6xHis_mVenus</b></p>====
| |
| - | <b>Investigator: Stefan</b><br>
| |
| - | <ul>
| |
| - | <b>Glycerol stocks were prepared:</b>
| |
| - | <li>B258 = pSB1C3_6xHis_middlelinker clone1</li>
| |
| - | <li>B259 = pSB1C3_6xHis_middlelinker clone2</li>
| |
| - | <br />
| |
| - |
| |
| - | <li>B260 = pSB1C3_6xHis_mVenus clone 1</li>
| |
| - | <li>B261 = pSB1C3_6xHis_mVenus clone 2</li>
| |
| - | <br />
| |
| - |
| |
| - | <b>Mini-Prep following the standard protocol</b>
| |
| - | <li>P310 = pSB1C3_6xHis_middlelinker clone1<br>
| |
| - | c= 185,31 ng/µl</li>
| |
| - | <li>P311 = pSB1C3_6xHis_middlelinker clone2<br>
| |
| - | c= 178,87 ng/µl</li>
| |
| - | <br />
| |
| - |
| |
| - | <li>P312 = pSB1C3_6xHis_mVenus clone 1<br>
| |
| - | c= 247,62 ng/µl</li>
| |
| - | <li>P313 = pSB1C3_6xHis_mVenus clone 2<br>
| |
| - | c= 268,98 ng/µl</li>
| |
| - | <br />
| |
| - | </ul>
| |
| - | <b>Test digestion</b><br>
| |
| - | <ul>
| |
| - | <li>Restriction-enzymes used: EcoRI and PstI</li>
| |
| - | </ul><br />
| |
| | {| border="1" | | {| border="1" |
| - | | align="left" | '''Components''' ||align="left"| '''P310+311''' ||align="left"| '''P312+313''' | + | | components || align="right" | V (pCMV_mVenus_YFP)/ µl ||align="right"| V(pCMV_MCS) / µl |
| | |- | | |- |
| - | | align="left" | DNA ||align="left"| 2 ||align="left"| 2
| + | | DNA || align="right" | 4,2||align="right"|3,6 |
| | |- | | |- |
| - | | align="left" | BSA (10x) ||align="left"| 1 ||align="left"| 1
| + | | BSA (10x) || align="right" |2||align="right"|2 |
| | |- | | |- |
| - | | align="left" | Buffer 4 (10x) ||align="left"| 1 ||align="left"| 1
| + | | Buffer 3 (10x)|| align="right" |2||align="right"|2 |
| | |- | | |- |
| - | | align="left" | EcoRI ||align="left"| 0,5 ||align="left"| 0,5 | + | |Enzyme: PstI/NotI HF (no.Lab:48/159)|| align="right" |1||align="right"|2 |
| | |- | | |- |
| - | | align="left" | PstI ||align="left"| 0,5 ||align="left"| 0,5 | + | |Enzyme: MluI (no.Lab:40)|| align="right" |1||align="right"|- |
| | |- | | |- |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 5 ||align="left"| 5 | + | |H2O|| align="right" |9,8||align="right"|10,4 |
| | |- | | |- |
| - | | align="left" | <b>Total volume</b> ||align="left"| <b>10</b> ||align="left"| <b>10</b> | + | |'''Total volume'''|| align="right" |<b>20</b>||align="right"|<b>20</b> |
| | |} | | |} |
| - | <br />
| |
| - | Incubation time: 90 minutes; Incubation temperature: 37°
| |
| - | <br />
| |
| - | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , running time: 55 minutes at 120 Volt <br />
| |
| - | <br />
| |
| - | <br />
| |
| - | [[File:Freiburg10 test digestion His middle+His mVenus.jpg|500px]]
| |
| - | <br />
| |
| - | <br />
| |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Test digestion looks good. Clones 1 of each plasmid (P310 and p312) were sent in for sequencing. </p>
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Repetition of test digestion of pAAV_RC_ins-rep-cap, pAAV_RC_ins-rep, pSB1C3_Rep40_RC_Insert and pSB1C3_Rep68_RC_Insert.</b></p>====
| |
| - | <b>Investigator: Chris L.</b><br>
| |
| - | <ul>
| |
| - | <li>P250 = pAAV_RC_ins-rep clone1<br>c=467,50 ng/µl</li>
| |
| - | <li>P314 = pAAV_RC_ins-rep-cap clone1<br>c=517,04 ng/µl</li>
| |
| - | <li>P315 = pAAV_RC_ins-rep-cap clone2<br>c=444,33 ng/µl</li>
| |
| - | <br />
| |
| - | <li>P316 = pSB1C3_Rep40_ins clone 1<br>c=302,22 ng/µl</li>
| |
| - | <li>P317 = pSB1C3_Rep40_ins clone 2<br>c=274,37 ng/µl</li>
| |
| - | <li>P318 = pSB1C3_Rep68_ins clone 1<br>c=491,69 ng/µl</li>
| |
| - | <li>P319 = pSB1C3_Rep68_ins clone 2<br>c=498,71 ng/µl</li>
| |
| - | </ul>
| |
| | <br> | | <br> |
| | + | <li> Incubation: 1 h at 37°C |
| | | | |
| - | <b>Test digestion:</b>
| |
| | <br> | | <br> |
| - | {| border="1"
| + | 1% Agarose gel |
| - | | components ||volume of '''P250'''/µl ||volume of '''P314'''/µl ||volume of '''P315'''/µl ||volume of '''P316'''/µl ||volume of '''P317'''/µl ||volume of '''P318'''/µl ||volume of '''P319'''/µl
| + | |
| - | |-
| + | |
| - | | DNA || 1 || 1 || 1 || 1 || 1 || 1 || 1
| + | |
| - | |-
| + | |
| - | | BSA (10x) ||1 ||1 || 1 || 1 || 1 || 1 ||1
| + | |
| - | |-
| + | |
| - | | Buffer 2 (10x) ||1 ||1 || 1 || 1 || 1 || 1 || 1
| + | |
| - | |-
| + | |
| - | |Enzyme Acc65I ||0,75 ||0,75 ||0,75 || - || - || - || -
| + | |
| - | |-
| + | |
| - | |Enzyme XcmI ||0,5 ||0,5 ||0,5 || - || - || - || -
| + | |
| - | |-
| + | |
| - | |Enzyme HindIII || - || - || - ||0,5 ||0,5 ||0,5 ||0,5
| + | |
| - | |-
| + | |
| - | |Enzyme BstEII || - || - || - ||0,5 ||0,5 ||0,5 ||0,5
| + | |
| - | |-
| + | |
| - | |H2O ||5,75 ||5,75 ||5,75 || 6 || 6 || 6 || 6
| + | |
| - | |-
| + | |
| - | |'''Total volume /µl'''||10 ||10 ||10 ||10 ||10 ||10 ||10
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | Incubation time: 1 h, Incubation temperature: 37° for P250, P314 and P315
| + | |
| - | <br />
| + | |
| - | Incubation time: 45 min, Incubation temperature: 37° with HindIII for P316, P317, P318 and P319 ; 45 min, Incubation temperature: 60° with BstEII for P316, P317, P318 and P319
| + | |
| - | >br />
| + | |
| - | Preparation of gel:<br />
| + | |
| - | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 120 Volt, running time: 45 minutes
| + | |
| - | <br />
| + | |
| - | {| align=right
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | *Marker: GeneRuler ladder mix
| + | |
| - | {| border="1"
| + | |
| - | |
| + | |
| - | !Marker
| + | |
| - | !Sample P250 10 /µl
| + | |
| - | !Sample P314 10 /µl
| + | |
| - | !Sample P315 10 /µl
| + | |
| - | !Sample P316 10 /µl
| + | |
| - | !Sample P317 10 /µl
| + | |
| - | !Sample P318 10 /µl
| + | |
| - | !Sample P319 10 /µl
| + | |
| - | |-
| + | |
| - | !Lane
| + | |
| - | |1
| + | |
| - | |3
| + | |
| - | |4
| + | |
| - | |5
| + | |
| - | |7
| + | |
| - | |8
| + | |
| - | |9
| + | |
| - | |10
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | [[File:Freiburg10 pAAV RC ins-rep-cap pSB1C3 Rep40 ins pSB1C3 Rep68 ins.png]]
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: The P314 and P315 looks like the control vector. The RepCap Vector_SDM_InsPvuII insertion didn´t work. The P316, P317, P318 and P318 was cut with the "wrong" enzymes. The same fragment size as the original vector appeared. Next time cut with EcoRI. </p>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Midi-Prep of pSB1C3_VCK_Bla</b></p>====
| + | |
| - | | + | |
| - | '''Investigators: Chris W. <br>
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Midi-Preps of P 320, B257</p>
| + | |
| - | The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | plasmid-no. || align="right" |P320
| + | |
| - | |-
| + | |
| - | | concentration (ng/µl)|| align="right" |408
| + | |
| - | |-
| + | |
| - | |}
| + | |
| | <br> | | <br> |
| - | | + | <li>1% agarose gel was repared, gel ran for 45 minutes(119 V) |
| - | ===103. labday 28.08.2010===
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Prep of pSB1C3_NLS</b></p>====
| + | |
| - | <b>Investigator: Stefan</b><br>
| + | |
| - | comments: No glycerol stock and test digestion was performed because bacteria culture inocculated from glyerol stock (B233).
| + | |
| - | <ul>
| + | |
| - | <b>Mini-Prep following the standard protocol</b>
| + | |
| - | <li>P321 = pSB1C3_NLS (1)<br> | + | |
| - | c= 180,45 ng/µl</li>
| + | |
| - | <li>P322 = pSB1C3_NLS (2)<br>
| + | |
| - | c= 187,26 ng/µl</li>
| + | |
| - | <br />
| + | |
| - | </ul>
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#cc0033;"><I><b>COMMENT (Patrick):</b> These mini-preps will be thrown away because B233 was used for inoculation due to a wrong entry in our plasmid excel sheet. B233 this is NOT the sequenced clone ! We sequenced B234/P282. Therefore 2x10 ml DYT + Cm were inoculated with B234. </i> </p>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cell culture</b></p>====
| + | |
| - | =====Harvest viral Stock and Transductiot of HT and A431 cells=====
| + | |
| - | <ol>
| + | |
| - | <li>P169: 100.000 cells</li>
| + | |
| - | <li>P169: 200.000 cells</li>
| + | |
| - | <li>P169: 300.000 cells</li>
| + | |
| - | <li>P169: 400.000 cells</li>
| + | |
| - | <li>P169: 500.000 cells</li>
| + | |
| - | <li>P169: 600.000 cells</li>
| + | |
| - | <li>P169: 700.000 cells</li>
| + | |
| - | <li>P262: 100.000 cells</li>
| + | |
| - | <li>P262: 200.000 cells</li>
| + | |
| - | <li>P262: 300.000 cells</li>
| + | |
| - | <li>P262: 400.000 cells</li>
| + | |
| - | <li>P262: 500.000 cells</li>
| + | |
| - | <li>P262: 600.000 cells</li>
| + | |
| - | <li>P262: 700.000 cells</li>
| + | |
| - | </ol>
| + | |
| - | <br />
| + | |
| - | 200.000 cells per well<br />
| + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>1</td>
| + | |
| - | <td>2</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>1</td>
| + | |
| - | <td>2</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>3</td>
| + | |
| - | <td>4</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>3</td>
| + | |
| - | <td>4</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>5</td>
| + | |
| - | <td>6</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>5</td>
| + | |
| - | <td>6</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>7</td>
| + | |
| - | <td>8</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>control, no virus</td>
| + | |
| - | <td>7</td>
| + | |
| - | <td>9</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" >
| + | |
| - | <tr>
| + | |
| - | <th width="80"> </th>
| + | |
| - | <th width="80">1</th>
| + | |
| - | <th width="80">2</th>
| + | |
| - | <th width="80">3</th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>A</td>
| + | |
| - | <td>control, no cells</td>
| + | |
| - | <td>11</td>
| + | |
| - | <td>13</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td>B</td>
| + | |
| - | <td>10</td>
| + | |
| - | <td>12</td>
| + | |
| - | <td>14</td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | =====Seeding AAV293 for Transfection at 30.8=====
| + | |
| - | <br />
| + | |
| - | 8 plates with 200.000 cells each were seeded
| + | |
| - | <br />
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Picking clones from the ViralBricks</b></p>====
| + | |
| - | Investigator: Volker
| + | |
| - | | + | |
| - | Clones from all 14 transformations of the ViralBricks were picked. Because the probability that the ligation worked and that the synthesized oligos do not conatin mutations is quite bad I picked 8 to 10 clones from each ligation, resulting in approximately 150 clones at all. The clones were used to inoculate 5ml of a mixture of 60%LB-media nd 40%DYP-media because our stocks were not sufficient for this experiment.
| + | |
| - | | + | |
| - | ===104. labday 29.08.2010===
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>P282/B234 mini-prep</b></p>====
| + | |
| - | ... was performed according to the standard protocol. <br>
| + | |
| - | Labelling:
| + | |
| - | * P323, pSB1C3_NLS clone 2.1, 129 ng/µl
| + | |
| - | * P324, pSB1C3_NLS clone 2.2, 141 ng/µl
| + | |
| - | Investigator: Patrick
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Digestion of pSB1C3_NLS (P324)</b></p>====
| + | |
| - | | + | |
| - | The NLS was cut out with NgoMIV and EcoRI, therefore the expected size of the hoped-for fragment is 55 bp. About 30 ng DNA can be detected in a 1% gel. According to my calculation there will be maximal 80 ng insert. <br>
| + | |
| - | 55bp/2019bp = 0,027 <br>
| + | |
| - | 0,027x140 ng/µl = 3,81 ng/µl <br>
| + | |
| - | 80 ng/3,81 ng/µl = 21 µl <br>
| + | |
| | <br> | | <br> |
| - | Digestion: 21 µl pSB1C3_NLS (P324), 2,5 µl EcoRI, 2,5 µl NgoMIV, 3 µl Buffer 4, 1 µl H2O. <br>
| + | Results: Test digestion of pCMV_MCS with NotI delivered plausibel results (expected fragment size: ~ 27 kbp and 17 kbp). |
| - | Digestion Time: 2 h 20 minutes.
| + | Unfortunately the digestion of pCMV_mVenus_YFP with PstI and MluI resulted in one 2.9 kbp and one 1.2 kbp fragment, which didn't correspond to the expected fragment sizes of 19 kbp and 33 kbp - but to fragment sizes of pCMV_MCS without mVenus_YFP! |
| - | | + | |
| - | The 2% agarose gel was loaded with 36 µl digesteion product including 6 µl 30% glycerol solution. I did not use loading dye because i to prevent that the dye overlay the mutual 55 bp insert.
| + | |
| - | | + | |
| - | [[File:Pat2908.JPG|500px]]
| + | |
| - | | + | |
| - | The gelextraction was performed according to the the standard protocl. The resolution of the picture ist not sufficient to be able to see the band clearly as the gel was put onto the UV-light table a very thin band with the expected size could be seen. <br>
| + | |
| - | DNA concentration measurement:
| + | |
| - | 3,28 & 7,7 ng/µl (two measurements). Especially the second value is doubtable because according to my calculation above the insert conccentration cant be higher than 3,81 ng/µl. Labeled: NLS, put into 4°C room.
| + | |
| - | | + | |
| - | New digestion over night following a gelrun tomorrow: 23 µl pSB1C3_NLS (P324), 2 µl EcoRI, 2 µl NgoMIV, 3 µl Buffer 4. <br>
| + | |
| - | | + | |
| - | Investigator: Patrick
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Harvesting cultures from the ViralBricks</b></p>====
| + | |
| - | Investigator: Volker
| + | |
| - | | + | |
| - | All approximately 150 cultures were pelletted (twice 2 ml) and the supernatant thrown away. The pellets were freezed and the rest of the culture stored in the 4°C room. These pellets will be preped, test digested and sequenced subsequently until right clones are identified.
| + | |
| - | | + | |
| - | ===105. labday 30.08.2010===
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>BioBrick assembly of pSB1C3_leftITR_pTERT & pSB1C3_ß-globin_YFP_hGH_rITR</b></p>====
| + | |
| - | | + | |
| - | Investigator: Achim <br />
| + | |
| - | | + | |
| - | comment: final assembly of a vector containing all aav elements & the tert promoter <br />
| + | |
| - | <b>Digestion:</b> <br />
| + | |
| - | {| border="1"
| + | |
| - | | components: || align="right" | pSB1C3_ß-globin_YFP_hGH_rITR(Vector): P294 || align="right" |pSB1C3_leftITR_pTERT (Insert): P256
| + | |
| - | |-
| + | |
| - | | DNA || align="right" |2,4 || align="right" |8,4
| + | |
| - | |-
| + | |
| - | | BSA (10x) || align="right" |2 || align="right" | 2
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x)|| align="right" |2 || align="right" |2
| + | |
| - | |-
| + | |
| - | |Enzyme1 || align="right" |1 (EcoRI) || align="right" |1 (EcoRI)
| + | |
| - | |-
| + | |
| - | |Enzyme2 || align="right" |1 (XbaI)|| align="right" |1 (SpeI)
| + | |
| - | |-
| + | |
| - | |H2O|| align="right" |11,6 || align="right" |5,6
| + | |
| - | |-
| + | |
| - | |'''Total volume'''|| align="right" | 20|| align="right" |20
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | | + | |
| - | | + | |
| - | Preparation of gel:<br />
| + | |
| - | 0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 110 Volt, running time: 45 minutes
| + | |
| - | <br />
| + | |
| - | | + | |
| - | | + | |
| - | [[Image:Freiburg10 30082010achim2.JPG|200px]] <br />
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | Expected sizes of constructs:
| + | |
| - | *pSB1C3_ß-globin_YFP_hGH_rITR: ~4000 bp
| + | |
| - | *pSB1C3_leftITR_pTERT: 2896 bp, ~640 bp
| + | |
| - | | + | |
| - | The corresponding bands were cut out and Gel-Extraction was performed according to protocol.
| + | |
| - | <br />
| + | |
| - | | + | |
| - | concentrations measured via NanoDrop:
| + | |
| - | *pSB1C3_ß-globin_YFP_hGH_rITR: 34.73 ng/µl
| + | |
| - | *pSB1C3_leftITR_pTERT: 9.48 ng/µl
| + | |
| - | | + | |
| - | | + | |
| - | <b>Ligation:</b> <br />
| + | |
| - | | + | |
| - | T4 ligation was performed according to protocol. <br />
| + | |
| - | | + | |
| - | Volumes used:
| + | |
| - | *vector: 2,9 µl
| + | |
| - | *insert: 5,1 µl
| + | |
| - | | + | |
| - | | + | |
| - | <b>Transformation:</b> <br />
| + | |
| - | | + | |
| - | bacterial strain used: XL1bB<br />
| + | |
| - | | + | |
| - | antibiotic used for plate: chlorampenicol<br />
| + | |
| - | | + | |
| - | Transformation was performed according to protocol.<br />
| + | |
| - | Plate was prepared and put in 37°C room.
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Continuation: Digestion of pSB1C3_NLS (P324)</b></p>====
| + | |
| - | | + | |
| - | Investigator: Patrick
| + | |
| - | | + | |
| - | [[File:Freigem10_Pat_3008.JPG|500px]]
| + | |
| - | | + | |
| - | The gelextraction was performed with QIAEX II Gel Extraction Kit which should also allow to extract fragments smaller than 70 bp (at least 40 bp). According to the QIAGEN Gel Extraction Kit the lenght of the DNA fragment should be about 70 bp.
| + | |
| - | Unfortunately the DNA fragment was blurred so a quite large piece had to be cut out.
| + | |
| - | | + | |
| - | The extraction yielded 4,26 and 3,77 ng/µl (two measurements) but this sample also quite polluted. ... put into freezer
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>CD biobrick production</b></p>====
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <b>Investigator: Kira</b><br>
| + | |
| - | NA samples were diluted 1:100
| + | |
| - | | + | |
| | <br> | | <br> |
| - | {| border="1"
| |
| - | | '''Ingredients''' || align="right" |CD sample
| |
| - | |-
| |
| - | | 5X Phusion HF buffer || align="right" |10 µl
| |
| - | |-
| |
| - | | 10 mM dNTP mix|| align="right" |1µl
| |
| - | |-
| |
| - | | forward primer: O158 || align="right" |2,5µl
| |
| - | |-
| |
| - | | reverse primer: O159 || align="right" |2,5 µl
| |
| - | |-
| |
| - | | DNA Template|| align="right" |0,5 µl
| |
| - | |-
| |
| - | | DMSO || align="right" |0 µl
| |
| - | |-
| |
| - | | Phusion Polymerase|| align="right" |0,5 µl
| |
| - | |-
| |
| - | |H<sub>2</sub>O|| align="right" |33µl
| |
| - | |-
| |
| - | |Total volume|| align="right" |50 µl
| |
| - | |}
| |
| | <br> | | <br> |
| - |
| |
| - | PCR program:
| |
| - |
| |
| - | {| border="1"
| |
| - | |Cycles||Temperature||Time
| |
| - | |-
| |
| - | |||98°C||1
| |
| - | |-
| |
| - | |8x||98°C||15"
| |
| - | |-
| |
| - | |||60°C||25"
| |
| - | |-
| |
| - | |||72°C||60"
| |
| - | |-
| |
| - | |17x||98°C||15"
| |
| - | |-
| |
| - | |||65°C||25"
| |
| - | |-
| |
| - | |||72°C||60"
| |
| - | |-
| |
| - | |1x||72°C||5'
| |
| - | |-
| |
| - | |Hold 4°C
| |
| - | |}
| |
| | <br> | | <br> |
| | + | <li>'''Ordering oligos:'''(Sigma-Aldrich) <br> |
| | + | (Bea)<br> |
| | + | Oligos have been ordered for modifying the ITRs. Deletion of PstI restriction site in ITR. <br> |
| | | | |
| - | Digestion of plasmid backbone:
| |
| - |
| |
| - | c (pSB1C3) = 151, 1 ng/ µl
| |
| - |
| |
| - | {| border="1"
| |
| - | | align="left" | '''Components''' ||align="left"| <b>vector</b> Volume/µL
| |
| - | |-
| |
| - | | align="left" | DNA 1 µg ||align="left"| 6,0 µl
| |
| - | |-
| |
| - | | align="left" | BSA (100x) ||align="left"| 0,2 µl
| |
| - | |-
| |
| - | | align="left" | Buffer no. 4 (10x) ||align="left"| 2,0 µl
| |
| - | |-
| |
| - | | align="left" | Enzyme 1 XbaI ||align="left"| 0,5 µl
| |
| - | |-
| |
| - | | align="left" | Enzyme 2 AgeI HF ||align="left"| 0,5 µl
| |
| - | |-
| |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 10,8 µl
| |
| - | |-
| |
| - | | align="left" | '''Total volume''' ||align="left"| <b>20</b>
| |
| - | |}
| |
| - | <br />
| |
| - |
| |
| - | incubation @ 37 C for approx. 2 h
| |
| - |
| |
| - |
| |
| - | Digestion of PCR product:
| |
| - | {| border="1"
| |
| - | | align="left" | '''Components''' ||align="left"| <b>PCR product</b> Volume/µL
| |
| - | |-
| |
| - | | align="left" | DNA ||align="left"| 30,0 µl
| |
| - | |-
| |
| - | | align="left" | BSA (100x) ||align="left"| 0,4 µl
| |
| - | |-
| |
| - | | align="left" | Buffer no. 4 (10x) ||align="left"| 4,0 µl
| |
| - | |-
| |
| - | | align="left" | Enzyme 1 XbaI ||align="left"| 1,5 µl
| |
| - | |-
| |
| - | | align="left" | Enzyme 2 AgeI HF ||align="left"| 1,0 µl
| |
| - | |-
| |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 3,1 µl
| |
| - | |-
| |
| - | | align="left" | '''Total volume''' ||align="left"| <b>40</b>
| |
| - | |}
| |
| - | <br />
| |
| - | incubation @ 37 C for approx. 2 h
| |
| - |
| |
| - | 1% agarose gel <br />
| |
| - |
| |
| - | Ligation <br />
| |
| - | T4 ligase was used <br />
| |
| - | 1 ul T4 Buffer <br />
| |
| - | 1 ul T4 Ligase <br />
| |
| - | 9 ul (6,9 ul vector+ 1,1 ul insert) DNA-mix <br />
| |
| - |
| |
| - | incubation over-night @ 18C <br />
| |
| - |
| |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Repetition of test digestion of pSB1C3_Rep40_RC_Insert and pSB1C3_Rep68_RC_Insert.</b></p>====
| |
| - | <b>Investigator: Chris L.</b><br>
| |
| - | <ul>
| |
| - | <li>P316 = pSB1C3_Rep40_ins clone 1<br>c=302,22 ng/µl</li>
| |
| - | <li>P317 = pSB1C3_Rep40_ins clone 2<br>c=274,37 ng/µl</li>
| |
| - | <li>P318 = pSB1C3_Rep68_ins clone 1<br>c=491,69 ng/µl</li>
| |
| - | <li>P319 = pSB1C3_Rep68_ins clone 2<br>c=498,71 ng/µl</li>
| |
| - | </ul>
| |
| | <br> | | <br> |
| - | <b>Test digestion:</b>
| + | '''right ITR of pAAV_MCS''' <br> |
| - | <br>
| + | |
| - | {| border="1"
| + | |
| - | | components ||volume of '''P316'''/µl ||volume of '''P317'''/µl ||volume of '''P318'''/µl ||volume of '''P319'''/µl
| + | |
| - | |-
| + | |
| - | | DNA || 1 || 1 || 1 || 1
| + | |
| - | |-
| + | |
| - | | BSA (10x) ||1 ||1 || 1 || 1
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x) ||1 ||1 || 1 || 1
| + | |
| - | |-
| + | |
| - | |Enzyme XcmI ||0,5 ||0,5 ||0,5 || 0,5
| + | |
| - | |-
| + | |
| - | |H2O || 6,5 || 6,5 || 6,5 || 6,5
| + | |
| - | |-
| + | |
| - | |'''Total volume /µl'''||10 ||10 ||10 ||10
| + | |
| - | |}
| + | |
| - | <br /> | + | |
| - | Incubation time: 1 h, Incubation temperature: 37°
| + | |
| - | >br />
| + | |
| - | Preparation of gel:<br />
| + | |
| - | 0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 120 Volt, running time: 45 minutes
| + | |
| - | <br />
| + | |
| - | [[File:Freiburg10 pSB1C3 Rep40 ins and pSB1C3 Rep68 ins.png|600px]]
| + | |
| - | <br />
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Test digestion looks good. Clones 2 of each plasmid (P317 and P319) were sent in for sequencing with VR2 Primer. </p>
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Midi-Prep of pAAV_RC_insertparts clone1</b></p>====
| + | oligos ordered:<br> |
| - | | + | |
| - | '''Investigators: Chris W. <br>
| + | fwd: 5´- gcgcagctgcctgcaCGGGCGCCTGATGCGG -3´ 77 °C, 31 bp <br> |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Midi-Preps of B208</p>
| + | rev: 5´- CCGCATCAGGCGCCCGTGCAGGCAGCTGCGC -3´ <br> |
| - | The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | plasmid-no. || align="right" |P325|| align="right" |p326
| + | |
| - | |-
| + | |
| - | | concentration (ng/µl)|| align="right" |2481,04 || align="right" |2050,66
| + | |
| - | |-
| + | |
| - | |}
| + | |
| | <br> | | <br> |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>FACS analysis, Seeding HT1080 and AAV293</b></p>====
| + | '''leftITR of pAAV_MCS'''<br> |
| | | | |
| - | '''Investigators: Kerstin, Adrian
| + | oligos ordered:<br> |
| | + | |
| | + | fwd: 5´- CCTTTTGCTCACATGTCGTGCAGGCAGCTGCGCG -3´ 74 °C, 34 bp <br> |
| | + | rev: 5´- CGCGCAGCTGCCTGCACGACATGTGAGCAAAAGG -3´<br> |
| | + | <br> |
| | + | For further details see link <br> |
| | | | |
| - | *FACS analysis of Transduction from 28.08.2010
| + | http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/images/4/44/Freiburg10_Oligos_ITR_mutagenesis_for_pAAV_delete_PstI.pdf |
| - | *Seeding HT1080 cells for Transduction (31.08.2010): 5x 6-well plates, 270.000 cells per well
| + | </li> |
| - | *Seeding AAV293 cells for Transfection (01.09.2010): 10 plates, 300.000 cells per well
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Preps and test digestion of Loop insertion BioBricks</b></p>====
| + | |
| - | Investigator: Anna
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Two clones of each ViralBrick were prepared and test digested. </p>
| + | |
| - | | + | |
| - | <b>Mini-Preps:</b>
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | || align="right" |'''p331''' || align="right" |'''p335'''|| align="right" |'''p339'''|| align="right" |'''p342'''|| align="right" |'''p345'''
| + | |
| - | | + | |
| - | |-
| + | |
| - | |Construct|| align="right" |pSB1C3_587_KO_BAP_clone3.1 || align="right" | pSB1C3_587_RGD_clone5.1 || align="right" | pSB1C3_453_His_clone7.1|| align="right" | pSB1C3_587_His_clone8.3 ||align="right" |pSB1C3_587_KO_empty_clone10.1
| + | |
| - | |-
| + | |
| - | |DNA-Concentration / µl|| align="right" |65,08 || align="right" | 94,29|| align="right" | 73,49|| align="right" |69,05|| align="right" | 66,73
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | | + | |
| - | <b>Test digestion:</b>
| + | |
| - | | + | |
| - | [[File:Freiburg10_LoopInsertions_Sal_Bam.jpg|600px]]
| + | |
| - | | + | |
| - | 1% Agarose gel: 1 g Agarose, 100 ml TAE, 6 µl Gelred <br/>
| + | |
| - | Marker: Gene ruler ladder DNA mix, 5 µl<br/>
| + | |
| - | | + | |
| - | Used enzymes: SalI, BamHI (in general, SDS has to be added because of the strong binding of enzymes to DNA!)
| + | |
| - | <br/>
| + | |
| - | <br/>
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Test digestion had no significant results. In addition, one sample was forgotten, it was not clear which one (description makes no sense). Test digestion has to be repeated.</p>
| + | |
| - | <br/>
| + | |
| - | <br/>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Repetition of subcloning Cap-insert into pAAV_RC</b></p>====
| + | |
| - | Investigator: Stefan
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comments</b>: There are still problems with BspMI. It was assumed that longer digestion could improve the results, therefore digestion with BspMI will be performed overnight.<br />
| + | |
| - | <b>P.S: Welcome back Hanna!</b></p>
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | '''Digestion of vector and insert:'''
| + | |
| - | *Insert: pMA_RepCap Vector_SDM_InsPvuII clone 1 (P211)
| + | |
| - | *Vector: pAAV_RC_ins-rep clone1 (P250)<br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | A two-step digestion was performed: <br />
| + | |
| - | <br />
| + | |
| - | '''1st step'''
| + | |
| - | {| border="1"
| + | |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| + | |
| - | |-
| + | |
| - | |DNA|| align="right" |8,5 || align="right" | 2
| + | |
| - | |-
| + | |
| - | |buffer 3|| align="right" |4,6 || align="right" | 5
| + | |
| - | |-
| + | |
| - | | Enzyme BsiWI|| align="right" |1 || align="right" | 1
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |32,4 || align="right" | 38
| + | |
| - | |-
| + | |
| - | |total volume|| align="right" |46 || align="right" |46
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | '''2nd step'''
| + | |
| - | {| border="1"
| + | |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| + | |
| - | |-
| + | |
| - | |Mix obtained from step 1 || align="right" |46 || align="right" | 46
| + | |
| - | |-
| + | |
| - | | Enzyme BspMI|| align="right" |4 || align="right" | 4
| + | |
| - | |-
| + | |
| - | |total volume|| align="right" |50 || align="right" |50
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | Digestion will be performed overnight and continued tomorrow.
| + | |
| - | <br/> | + | |
| - | <br/>
| + | |
| - | ===106. labday 31.08.2010===
| + | |
| - | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Cloning ZEGFR:1907 and 6xHis_middlelinker into pCerulean</b></p>====
| + | |
| - | Investigator: Hanna
| + | |
| - | <br/>
| + | |
| - | <p><font color="#ff00ff"><b>Comment:</b> In order to fuse the His-Tag to VP2, the His-Tag-Middlelinker construct will be cloned into the pCerulean plasmid (contains CMV promoter and SV40 terminator) today. In the next step, VP2 will be cloned behind the His-Tag-Middlelinker.
| + | |
| - | <br/>
| + | |
| - | In addition to that the Affibody (ZEGFR:1907) will be also cloned into pCerulean (without linker). Also here, the next step will be fusing VP2 behind the this targeting molecule.</font></p>
| + | |
| - | <br/>
| + | |
| - | | + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;"><u>Practical Cloning:</u></p>
| + | |
| - | *plasmid:
| + | |
| - | **Vector: name: pCerulean; number: P273
| + | |
| - | **Insert: name: pSB1C3_6xHis_middlelinker (P310) <b>+</b> pSB1C3_Zegfr:1907 (because P267 was empty, P285 was used! See lab day 23.08.!)
| + | |
| - | *new vector name: pCerulean_6xHis_middlelinker <b>+</b> pCerulean_Zegfr:1907
| + | |
| - | *buffer used: 4; Restriction-enzymes used: XbaI and PstI
| + | |
| - | <br />
| + | |
| - | '''Comments:''' Because the P267 tube was empty (!), P285 was used which was prepared by Jessy - I don't know whether this is a trafo of the glycerol stock? Sequenced?
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Digestion</p>
| + | |
| - | {| border="1"
| + | |
| - | | components || align="right" |volume of pCerulean/µl || align="right" |volume of pSB1C3_6xHis_middlelinker/µl || align="right" |volume of pSB1C3_ZEGFR:1907
| + | |
| - | |-
| + | |
| - | | DNA || align="right" | 3.6 || align="right" | 16.2 || align="right" | 11.5
| + | |
| - | |-
| + | |
| - | | BSA (10x) || align="right" | 3 || align="right" | 2.5 || align="right" | 2
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x)|| align="right" | 3 || align="right" | 2.5 || align="right" | 2
| + | |
| - | |-
| + | |
| - | |Enzyme 1 XbaI || align="right" | 1.5 || align="right" | 1 || align="right" | 1
| + | |
| - | |-
| + | |
| - | |Enzyme 2 PstI || align="right" | 1.5 || align="right" | 1 || align="right" | 1
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" | 17.4 || align="right" | 1.8 || align="right" | 2.5
| + | |
| - | |-
| + | |
| - | |'''Total volume'''|| align="right" | 30 || align="right" | 25 || align="right" | 20
| + | |
| - | |}
| + | |
| - | *Incubation: 1.5 h
| + | |
| - | <br /><br />
| + | |
| - | | + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Agarose-Gel:</p>
| + | |
| - | <br />
| + | |
| - | 0.8 g Agarose, 55 ml TAE (1.45 %), 3 µL GELRED, at 115 Volt, running time: 35 minutes
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | {| border="1" cellspacing="0" cellpadding="2" bordercolor="black"
| + | |
| - | !Sample
| + | |
| - | !Sample/µl]
| + | |
| - | !Loading dye (6x)/µl
| + | |
| - | !Expected size 1 (Geneious)
| + | |
| - | |--
| + | |
| - | |P285
| + | |
| - | |20 µl
| + | |
| - | |4 µl
| + | |
| - | |210 bp
| + | |
| - | |--
| + | |
| - | |P273
| + | |
| - | |30 µl
| + | |
| - | |6 µl
| + | |
| - | |~ 3900 bp
| + | |
| - | |--
| + | |
| - | |P310
| + | |
| - | |25 µl
| + | |
| - | |5 µl
| + | |
| - | |86 bp
| + | |
| - | | + | |
| - | |}
| + | |
| - | {| align=right
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | *Marker: GeneRuler ladder mix
| + | |
| - | {| border="1"
| + | |
| - | |
| + | |
| - | !Marker /µL
| + | |
| - | !Sample P285 /µl
| + | |
| - | !Sample P273 /µl
| + | |
| - | !Sample P310 /µl
| + | |
| - | |-
| + | |
| - | !Lane
| + | |
| - | |5.5
| + | |
| - | |24
| + | |
| - | |25
| + | |
| - | |25
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | | + | |
| - | [[File:Freiburg10_Digestion_6xHis_middlelinker_and_Zegfr.JPG.gif|700px]]
| + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Gel extraction</p>
| + | |
| - | <br />
| + | |
| - | Gel measurement:
| + | |
| - | <br />
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Sample'''
| + | |
| - | | align="left" | '''Weight'''
| + | |
| - | | align="left" | '''Volume'''
| + | |
| - | | align="left" | '''Concentration'''
| + | |
| - | |-
| + | |
| - | | align="left" | P285
| + | |
| - | | align="left" | 240 mg
| + | |
| - | | align="left" | 20 µL
| + | |
| - | | align="left" | 5.4 ng/µL
| + | |
| - | |-
| + | |
| - | | align="left" | P273
| + | |
| - | | align="left" | 130 mg
| + | |
| - | | align="left" | 20 µL
| + | |
| - | | align="left" | 14.5 ng/µL
| + | |
| - | |-
| + | |
| - | | align="left" | P310
| + | |
| - | | align="left" | 220 mg
| + | |
| - | | align="left" | 20 µL
| + | |
| - | | align="left" | 2.4 ng/µL
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | <b>Comment:</b> The DNA concentrations were very bad, which could be due to the fact, that the 6xHis_middlelinker construct's size is just 86 bp and the affibody's size is just 210 bp...
| + | |
| - | <br />
| + | |
| - | | + | |
| - | | + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Ligation</p>
| + | |
| - | <br />
| + | |
| - | <b>1. pCerulean_6xHis_middlelinker</b>
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | ||align="left"| '''6xHis_middlelinker''' ||align="left"| '''pCerulean'''
| + | |
| - | |-
| + | |
| - | | align="left" | '''Volume/µl''' ||align="left"| 2.57 ||align="left"| 6.43
| + | |
| - | |}
| + | |
| - | <br/>
| + | |
| - | <b>2. pCerulean_Zegfr:1907</b>
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | ||align="left"| '''Zegfr:1907''' ||align="left"| '''pCerulean'''
| + | |
| - | |-
| + | |
| - | | align="left" | '''Volume/µl''' ||align="left"| 2.72 ||align="left"| 6.28
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | | + | |
| - | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Trafo</p>
| + | |
| - | <br />
| + | |
| - | Trafo was performed following the standard protocol. Cells: XL1b (60 µL), DNA amount per sample: 3 µL; cells were plated onto Kanamycin-plates.
| + | |
| - | <br/>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>PCR of SV 40 terminator</b></p>====
| + | |
| - | <b>Investigator: Bea</b>
| + | |
| - | <br/>
| + | |
| - | <p style="color:#66bbff;"><b>Comment:</b> In order to obtain the SV40 poly adenylation site and clone it into the iGEM standard plasmid pSB1C3 a PCR needs to be performed with the pEGFP-C1 which contains the SV40 terminator.</p>
| + | |
| - | <br/>
| + | |
| - | <ul>
| + | |
| - | <li>Plasmid used for PCR: pEGFP-C1 (P230)</li>
| + | |
| - | <li>1:1000 dilution of P230 has been prepared</li>
| + | |
| - | <li>Plasmid used as vector: pSB1C3_CFP1 (P51.2)</li>
| + | |
| - | </ul>
| + | |
| - | <br/>
| + | |
| - | <b>PCR (was performed following the standard protocol)</b>
| + | |
| | <br> | | <br> |
| | | | |
| - | {| border="1"
| + | '''Test-Trafo of chemical competent E.coli cells''' <br> |
| - | | '''Ingredients''' || align="right" |'''Volume / µl'''
| + | (Hanna) |
| - | |-
| + | <br> |
| - | | 5X Phusion HF buffer || align="right" |10
| + | The competent E.coli XL-1-blue and competent E.coli BL21 which were prepared the day before were test-transformed with pUC18 - following the standard protocol. Cells were plated on agar plates containing ampicillin and stored over night at 37°C. |
| - | |-
| + | Further on two control plates containing ampicillin or kanamycin were prepared. Non-transformed cells were plated on them and also stored over night at 37°C. |
| - | | 10 mM dNTP mix|| align="right" |1
| + | |
| - | |-
| + | |
| - | | forward primer: O160 || align="right" |2,5
| + | |
| - | |-
| + | |
| - | | reverse primer: O161|| align="right" |2,5
| + | |
| - | |-
| + | |
| - | | DNA Template|| align="right" |3
| + | |
| - | |-
| + | |
| - | |- DMSO || align="right" |-
| + | |
| - | |-
| + | |
| - | | Phusion Polymerase|| align="right" |0,5
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |30,5
| + | |
| - | |-
| + | |
| - | |'''Total volume'''|| align="right" |'''50'''
| + | |
| - | |}
| + | |
| | <br> | | <br> |
| - |
| |
| - | <b>PCR program:</b>
| |
| - | {| border="1"
| |
| - | |'''PCR Program'''|| align="right" |'''temperature/ °C'''|| align="right" |'''Time'''
| |
| - | |-
| |
| - | |1|| align="right" |98 || align="right" |1min
| |
| - | |-
| |
| - | |2|| align="right" |98 || align="right" |15s
| |
| - | |-
| |
| - | |8x|| align="right" |59 || align="right" |25s
| |
| - | |-
| |
| - | |3 || align="right" |72|| align="right" |7s
| |
| - | |-
| |
| - | |4 || align="right" |98|| align="right" | 15s
| |
| - | |-
| |
| - | |17x|| align="right" |67|| align="right" |25s
| |
| - | |-
| |
| - | |5|| align="right" |72|| align="right" |7s
| |
| - | |-
| |
| - | |6x|| align="right" |72|| align="right" |5min
| |
| - | |-
| |
| - | |Hold|| align="right" |4|| align="right" |
| |
| - | |}
| |
| - | After the PCR was performed, the sample was loaded on a 1.5% agarose gel and run for 30 minutes. As it can be seen in the picture below the expected size of the PCR product (260bp) can be confirmed. The lower band corresponds to the added primers and does not seem to be an additional band.
| |
| - |
| |
| - | <gallery widths=300px heights=300px>
| |
| - | Image:Freiburg10 PCR product SV40.jpg
| |
| - | </gallery>
| |
| - |
| |
| | <br> | | <br> |
| - | For cloning the obtained fragment into the pSB1C3 standard vector, the construct need to be digested with the same restriction enzymes as the PCR product will be cut with. In this case the primers were designed with XbaI overhang for the prefix, and with SpeI-NotI-PstI overhang for the suffix. Therefore the vector will be cut with XbaI and PstI. The sample were incubated at 37 C for approx. 1.5 h
| |
| - | <br />
| |
| - | <ul>
| |
| - | <li>Plasmid used: pSB1C3_CFP (P51.2) c=150ng/µL</li>
| |
| - | <li>Add BSA</li>
| |
| - | <li>XbaI and PstI were used</li>
| |
| | </ul> | | </ul> |
| - | <br/>
| |
| - | <b>Digestion of vector:</b>
| |
| - |
| |
| - | {| border="1"
| |
| - | | align="left" | '''Components''' ||align="left"| <b>v<sub>pSB1C3_CFP</sub></b>/µL
| |
| - | |-
| |
| - | | align="left" | DNA ||align="left"| 8
| |
| - | |-
| |
| - | | align="left" | BSA (100x) ||align="left"| 2
| |
| - | |-
| |
| - | | align="left" | Buffer no. 4 (10x) ||align="left"| 2
| |
| - | |-
| |
| - | | align="left" | Enzyme 1 XbaI ||align="left"| 1
| |
| - | |-
| |
| - | | align="left" | Enzyme 2 AgeI HF ||align="left"| 1
| |
| - | |-
| |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 6
| |
| - | |-
| |
| - | | align="left" | '''Total volume''' ||align="left"| <b>20</b>
| |
| - | |}
| |
| - | <br />
| |
| - | <ul>
| |
| - | <li>Incubation of the sample at 37°C</li>
| |
| - | <li>Incubation for 120 minutes</li>
| |
| | </ul> | | </ul> |
| - | <br />
| + | ===16. Labortag 27.05.2010=== |
| - | After the PCR has been cut out of the gel and purified with the Qiagen Gel Extraction Kit obtaining a concentration of 45 ng/µl measured with the Nanodrop, the complete PCR product volume of 29 µL was digested with XbaI and PstI for 2 hours.<br />
| + | |
| - | <br />
| + | |
| - | <b>Digestion of the PCR product SV40</b>
| + | |
| - | {| border="1"
| + | |
| - | | align="left" | '''Components''' ||align="left"| <b>v<sub>PCR product</sub></b>/µL
| + | |
| - | |-
| + | |
| - | | align="left" | DNA ||align="left"| 29,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | BSA (10x) ||align="left"| 4,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | Buffer no. 4 (10x) ||align="left"| 4,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 1 XbaI ||align="left"| 1,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | Enzyme 2 AgeI HF ||align="left"| 1,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | H<sub>2</sub>O ||align="left"| 1,0 µl
| + | |
| - | |-
| + | |
| - | | align="left" | '''Total volume''' ||align="left"| <b>40</b>
| + | |
| - | |}
| + | |
| - | <ul>
| + | |
| - | <li>Incubation of the sample at 37°C</li>
| + | |
| - | <li>Incubation for 120 minutes</li>
| + | |
| - | </ul>
| + | |
| - | <br />
| + | |
| - | The digested PCR product SV40 was purifed with the PC Purification Kit and concentration was measured.
| + | |
| - | <ul>
| + | |
| - | <li><b>c<sub>SV40</sub>= 31,5 ng/µl</b></li>
| + | |
| - | <li><b>c<sub>pSB1C3</sub>= 7,74 ng/µL</b></li>
| + | |
| - | </ul>
| + | |
| - | For T4 ligation of the two fragments, the needed volumes of insert (SV40) and vector (pSB1C3) were calculated with the labtools program. The total volume DNA (vector/insert) mix will be 8 µL.
| + | |
| - | <ul>
| + | |
| - | <li><b>v<sub>SV40</sub>= 0,67 µL</b></li>
| + | |
| - | <li><b>v<sub>pSB1C3</sub>= 7,33 µL</b></li>
| + | |
| - | <li><b>v<sub>T4 buffer</sub>= 1 µL</b></li>
| + | |
| - | <li><b>v<sub>t4 ligase</sub>= 1 µL</b></li>
| + | |
| - | </ul>
| + | |
| - | The ligation reaction was carried out for 40 minutes at room temperature. The ligated plasmid was transformed into <b>XL1-Blue cells</b> and plated on agar plates containing the appropriate amount of <b>chloramphenicol</b>.
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#33FF88;"><b>Cloning of NLS into pCerulean_VP1up</b></p>==== | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Cell counting</b></p>==== |
| - | Investigator: Anissa
| + | |
| - | <br/>
| + | |
| - | <p style="color:#33FF88;"><b>Comment:</b> NLS was already 2 times cut of the pSB1C3_NLS and purified by Patrick (NLS 1: of the 29.08.10, NLS 2: of the 30.08.10) . Now, pCerulean_VP1up will be digested and NLS will be ligated into it.</p>
| + | |
| - | <br/>
| + | |
| - | <ul>
| + | |
| - | <li>Digestion of pCerulean_VP1up (p303)
| + | |
| - | <br/><br/>
| + | |
| - | {| border="1"
| + | |
| - | | components || align="right" |Vector
| + | |
| - | |-
| + | |
| - | | DNA || align="right" |2,6
| + | |
| - | |-
| + | |
| - | | BSA (10x) || align="right" |1,5
| + | |
| - | |-
| + | |
| - | | Buffer 4 (10x)|| align="right" |1,5
| + | |
| - | |-
| + | |
| - | |Enzyme PstI || align="right" |1
| + | |
| - | |-
| + | |
| - | |Enzyme AgeI || align="right" |1
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |7,4
| + | |
| - | |-
| + | |
| - | |'''Total volume '''|| align="right" |15
| + | |
| - | |}
| + | |
| - | <br>
| + | |
| - | </li>
| + | |
| - | <li>Gel:
| + | |
| - | <br/>
| + | |
| - | A 1% gel was made and the sample was running 30 minutes.<br/>
| + | |
| - | [[image:Anissa310810.png|300px]]<br/>
| + | |
| - | Afterwards a gelextraction according the qiagen-protocol was performed.<br/><br/>
| + | |
| - | </li>
| + | |
| - | <li>Ligation: <br/>
| + | |
| - | {| border="1"
| + | |
| - | | components || align="right" |Volume /µl || align="right" |concentration /ng/µl
| + | |
| - | |-
| + | |
| - | | NLS 2 insert of 30.08.10 || align="right" |1,51 || align="right" |3
| + | |
| - | |-
| + | |
| - | | NLS 1 insert of 29.08.10|| align="right" |1,51|| align="right" |3
| + | |
| - | |-
| + | |
| - | | pCerulean_VP1up|| align="right" |6,49 || align="right" |29
| + | |
| - | |}<br/>
| + | |
| - | </li><br/>
| + | |
| - | <li>Transfomation: was performed into XL1 blue on a kanamycin plate according the standard protocol.
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| | | | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)</b></p>====
| + | <p><b>Investigator: Patrick, Adrian, Chris W. Christian L.</b></p> |
| | | | |
| - | Investigator: Patrick <br>
| + | * Meeting |
| - | P276 digestion (207 ng/µl): 14 µl DNA sample, 2 µl Buffer 4, 2 µl BSA, 1 µl XbaI, 1 µl PstI-HF. <br>
| + | |
| - | P273 digestion (419 ng/µl): 4 µl DNA sample, 1 µl Buffer 4, 1 µl BSA, 1 µl XbaI, 1 µl PstI-HF. <br>
| + | |
| - | Digestion time: 1 h 45 min.
| + | |
| | | | |
| - | Expected size of the fragments: <br>
| + | cell counting: |
| - | P276: 786 & 2058 bp <br>
| + | <br> |
| - | P273: 756 & 3938 bp <br>
| + | * BL21 + PUC18 Trafo 54*4 = 216 |
| - | | + | * XL1B + PUC18 Trafo 30*4 = 120 |
| - | [[image:Cartoons48.jpg|thumb|right| My dear friend, where is your initial comment? Nice gel :)
| + | digitalization of recipe-cards (Christian L.) |
| - | Yours sincerely, Hanna :)]]
| + | |
| - | see: http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/August_2010#Strategy_for_VP2_N-terminal_fusion_and_VP1_insertion_of_targeting_moelcules
| + | |
| - | <br/>
| + | |
| - | <font color="#ff00ff">Haha, good idea! :) </font><br/>
| + | |
| - | [[File:Freiburg10_3108pat2.JPG|500px]]
| + | |
| - | | + | |
| - | | + | |
| - | Gelextraction:
| + | |
| - | Obviously there was something wrong with the gel or the sample. The marked bands were cut out and the gelextraction was performed acording to he standard protocol, yielding the following concentrations:<br>
| + | |
| - | P276 up: 12,6 ng/µl <br>
| + | |
| - | P276 down: 9,9 ng/µl <br>
| + | |
| - | P273: 37,7 ng/µl <br>
| + | |
| - | | + | |
| - | All of them were polluted.
| + | |
| - | | + | |
| - | There were two bands at a level where no bands should be. P 276 looks also strange because accidentally 6,5 µl 1kb GeneRuler were pipetted into the sample. The expected bands are all blurred. Even the expected 756 bp fragment of P273 is not at the level it should be. To ensure i dont throw away the insert i cut out both bands from P276. Now tomorrow there will be a ligation and a transformation with two samples instead of one
| + | |
| - | | + | |
| - | | + | |
| - | <br/>
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Sequencing results of pSB1C3_6xHis_middlelinker and pSB1C3_6xHis_mVenus</b></p>====
| + | |
| - | Investigator: Hanna
| + | |
| - | <br/>
| + | |
| - | | + | |
| - | <p><b>1st Comment:</b> Sequencing of pSB1C3_6xHis_middlelinker looked good:</p>
| + | |
| - | <br/>
| + | |
| - | [[File:Freiburg10 6xHis middlelinker.JPG|600px]]
| + | |
| - | <br/>
| + | |
| - | <p><b>2nd Comment:</b> Sequencing of mVenus looked good. The His-Tag delivered 2 additional bp and the suffix 1 transition (see picture). This could be due to bad sequencing qualities at this region... but needs to be verified!</p>
| + | |
| - | [[File:Freiburg10 6xHis mVenus.JPG|700px]]
| + | |
| - | | + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Contiuation: Repetition of subcloning Cap-insert into pAAV_RC</b></p>====
| + | |
| - | <b>Investigator: Stefan</b>
| + | |
| - | | + | |
| - | <b>Comments:</b> Overnight digestion does not look good, therefore a new approach will be performed today using different concentrations of spermidine (0,5mM, 2,5mM and 10mM) because in Oller et al (Biochemistry, 1991) it was suggested that this improves digestion with BspMI.
| + | |
| - | | + | |
| - | A two-step digestion was performed: <br />
| + | |
| - | <br />
| + | |
| - | '''1st step'''
| + | |
| - | {| border="1"
| + | |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| + | |
| - | |-
| + | |
| - | |DNA|| align="right" |8,5 || align="right" | 2
| + | |
| - | |-
| + | |
| - | |buffer 3|| align="right" |4,6 || align="right" | 5
| + | |
| - | |-
| + | |
| - | | Enzyme BsiWI|| align="right" |1 || align="right" | 1
| + | |
| - | |-
| + | |
| - | |H<sub>2</sub>O|| align="right" |32,4 || align="right" | 38
| + | |
| - | |-
| + | |
| - | |total volume|| align="right" |46 || align="right" |46
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | *Incubation: 70 minutes; 55°C<br> | + | |
| - | '''2nd step'''
| + | |
| - | {| border="1"
| + | |
| - | | || align="right" |'''P211 / µl''' ||'''P250 / µl'''
| + | |
| - | |-
| + | |
| - | |Mix obtained from step 1 || align="right" |46 || align="right" | 46
| + | |
| - | |-
| + | |
| - | | Enzyme BspMI|| align="right" |4 || align="right" | 4
| + | |
| - | |-
| + | |
| - | |total volume|| align="right" |50 || align="right" |50
| + | |
| - | |}
| + | |
| - | <br />
| + | |
| - | *Incubation: 2,5h ; 37°C<br> | + | |
| - | '''Agarosegel'''
| + | |
| - | <br />
| + | |
| - | 0.4 g Agarose, 50 ml TAE (0,8 %), 3 µL GELRED, at 115 Volt
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | | + | |
| - | | + | |
| - | <br />
| + | |
| - | [[File:Freiburg10 P250 without Spermidine.png|300px|left|thumb|]]
| + | |
| - | [[File:Freiburg10 P250 with spermidine.png|500px|right|thumb|]]
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | <br />
| + | |
| - | | + | |
| - | '''Gel extraction'''
| + | |
| - | <br />
| + | |
| - | Gel-Ex was performed according to protocol.<br />
| + | |
| - | NanoDrop:
| + | |
| - | *pMA_RepCap-insert: c= 7,42 ng/µl
| + | |
| - | *pAAV_RC_ins-rep: c=12,9 ng/µl
| + | |
| - | <br />
| + | |
| - | | + | |
| - | '''T4 Ligation'''
| + | |
| - | <br />
| + | |
| - | Volume insert: 3,43 µl<br>
| + | |
| - | Volume vector: 4,57 µl<br>
| + | |
| - | Ligation incubated for 50 minutes at room temperature.<br />
| + | |
| - | <br />
| + | |
| - | '''Transformation'''
| + | |
| - | Trafo was preformed according to standard protocol using XL1blue cells on plates containing Ampicilin.
| + | |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>2nd Repetition of test digestion of Loop insertion BioBricks</b></p>====
| + | |
| - | <b>Investigator: Achim, Anna </b><br>
| + | |
| - | | + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: First test digestion didn't work (see 31.08.10), this time a new approach was done with NotI. </p>
| + | |
| - | | + | |
| - | <b>Test digestion:</b>
| + | |
| | <br> | | <br> |
| - | {| border="1"
| |
| - | | components ||Volume for each sample /µl
| |
| - | |-
| |
| - | | DNA || 10
| |
| - | |-
| |
| - | | BSA (10x) ||1,5
| |
| - | |-
| |
| - | | Buffer 4 (10x) ||1,5
| |
| - | |-
| |
| - | |Enzyme NotI ||0,5
| |
| - | |-
| |
| - | |H2O || 1,5
| |
| - | |-
| |
| - | |'''Total volume /µl'''||15
| |
| - | |}
| |
| - | <br />
| |
| | | | |
| - | {| border="1"
| + | ===17. Labortag 31.05.2010=== |
| - | |align="left" | '''Sample-No.''' || align="left" | '''1+2''' ||align="left" | '''3+4''' ||align="left" | '''5+6''' ||align="left" | '''7+8''' ||align="left" | '''9+10''' ||align="left" | '''11+12''' ||align="left" | '''13+14''' ||align="left" | '''15+16''' ||align="left" | '''17+18''' ||align="left" | '''19+20''' ||align="left" | '''21-23''' ||align="left" | '''24+25''' ||align="left" | '''26+27''' ||align="left" | '''28+29''' ||align="left" | '''30''' ||align="left" | '''31'''
| + | |
| - | |-
| + | |
| - | |align="left" | '''Components''' ||align="left"| '''453 BAP''' ||align="left"| '''587 BAP''' ||align="left"| '''587 KO BAP'''||align="left"| '''453 RGD'''||align="left"| '''587 RGD'''||align="left"| '''587 KO RGD'''||align="left"| '''453 HIS'''||align="left"| '''587 HIS'''||align="left"| '''587 KO HIS'''||align="left"| '''587 KO EMPTY'''||align="left"| '''453 Z34C'''||align="left"| '''587 Z34C'''||align="left"| '''587 KO Z34C'''||align="left"| '''587 KO Z34C SPACER'''||align="left"| '''pSb1C3_Bla (cut version)'''||align="left"| '''pSb1C3_Bla (uncut)'''
| + | |
| - | |-
| + | |
| - | | align="left" | Oligo 1||align="left"|O124: 10 ||align="left"|O126: 10 ||align="left"|O128: 10 ||align="left"|O130: 10 ||align="left"|O132: 10 ||align="left"|O134: 10 ||align="left"|O135: 10 ||align="left"|O137: 10 ||align="left"|O139: 10 ||align="left"|O141: 10 ||align="left"|O143: 10 ||align="left"|O145: 10 ||align="left"|O147: 10 ||align="left"|O149: 10 ||align="left"|--- ||align="left"|---
| + | |
| - | |-
| + | |
| - | | align="left" | Oligo 2 ||align="left"|O125: 10 ||align="left"|O127: 10 ||align="left"|O129: 10 ||align="left"|O131: 10 ||align="left"|O133: 10 ||align="left"|O151: 10 ||align="left"|O136: 10 ||align="left"|O138: 10 ||align="left"|O140: 10 ||align="left"|O142: 10 ||align="left"|O144: 10 ||align="left"|O146: 10 ||align="left"|O148: 10 ||align="left"|O150: 10 ||align="left"|--- ||align="left"|---
| + | |
| - | |-
| + | |
| - | |}
| + | |
| | | | |
| - | <br/> | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>pAAV_RC R585A and R588A transistions</b></p>==== |
| - | Incubation time: 1 h, Incubation temperature: 37°
| + | |
| - | <br/> | + | |
| | | | |
| - | <b>Preparation of gel:</b><br/>
| |
| - | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
| |
| - | <br />
| |
| - | <br/>
| |
| - | [[File:Freiburg10_LoopInsertions.jpg|600px]]
| |
| - | <br />
| |
| | | | |
| - | <br/>
| |
| - | <b>Expected fragment sizes /bp:</b> <br/>
| |
| - | pSB1C3: 2051 bp
| |
| - | Bla: 905<br/>
| |
| - | BAP: ~150<br/>
| |
| - | RGD: ~ 130<br/>
| |
| - | Z34C: ~ 200<br/>
| |
| - | His: ~ 125 <br/>
| |
| - | <br/>
| |
| | | | |
| - | The following clones were sent for sequencing: 2,3,5,7,9,11,13,16,17,19 and 25.
| + | In order to alter the tropism of AAV2 several modifications have to be performed. |
| - | <br />
| + | First of all the binding to Heparan Sulfate Proteoglycan has to be disrupted. This can be done by R585A and R588A transistions: |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Clones (see above) were sent for sequencing with VR2 Primer. In addition, 8 mL DYT medium was inoculated with each of the clones for preparation of glycerol stocks. For sequencing results go to labday 01.09.10.</p>
| + | [[File:Freiburg10 R585A R588A.jpg|800px|]] |
| - | | + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Midi-Prep of pAAV_RC & pHelper</b></p>====
| + | |
| - | | + | |
| - | '''Investigators: Chris W. <br>
| + | |
| - | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Midi-Preps of pHelper=P356 and pAAV_RC=P357</p>
| + | |
| - | The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | plasmid-no. || align="right" |P356|| align="right" |P357
| + | |
| - | |-
| + | |
| - | | concentration (ng/µl)|| align="right" |1068,0 || align="right" |1274.80
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | <br>
| + | |
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b>Transformation of Biobrick CD</b></p>====
| + | |
| - | <b>Investigator: Kira</b>
| + | |
| - | <br/>
| + | |
| - | Transformation was performed according to the standard protocol.
| + | |
| | | | |
| | <html> | | <html> |
| | </div> | | </div> |
| | </html> | | </html> |

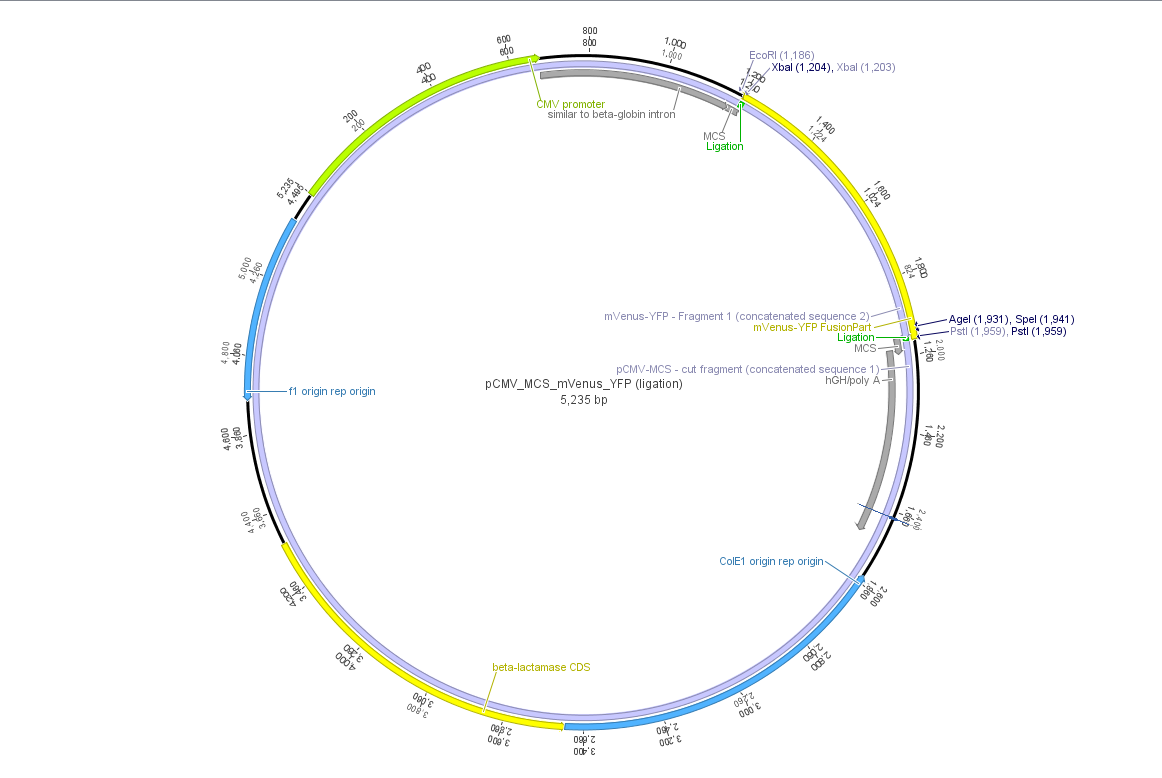

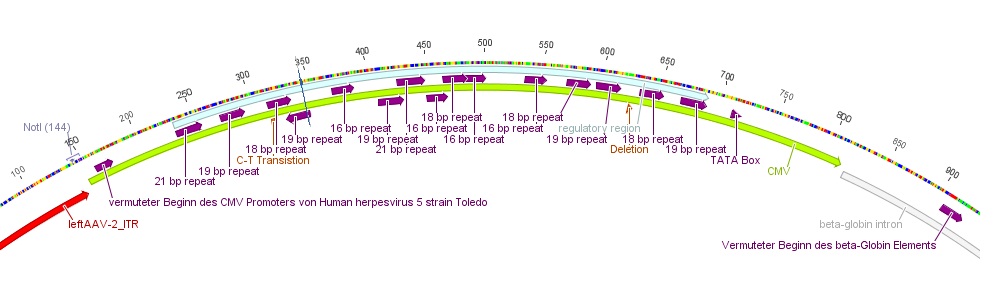
 "
"