Team:Freiburg Bioware/testpage
From 2010.igem.org
| Line 122: | Line 122: | ||
<div class="box_right"> | <div class="box_right"> | ||
</html> | </html> | ||
| - | ==== | + | ==September== |
| - | <br> | + | ===107. labday 01.09.2010=== |
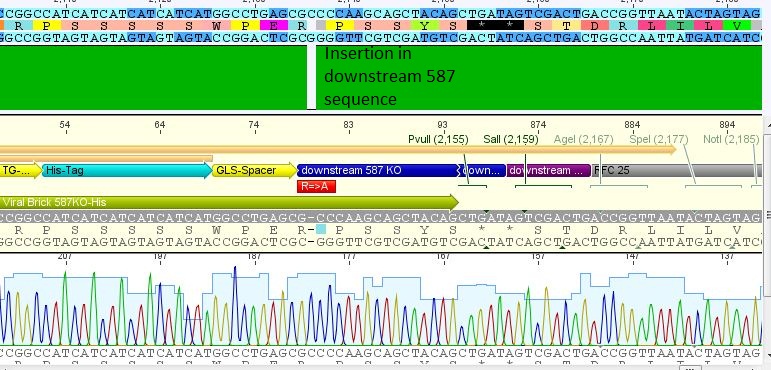
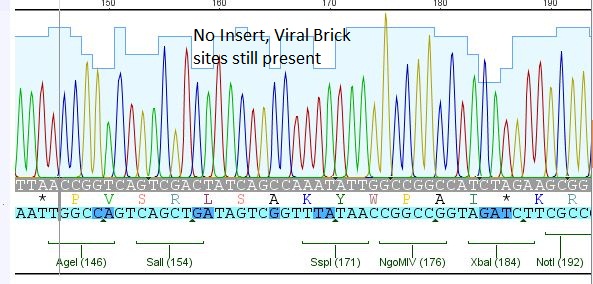
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Loop insertion BioBricks: Sequencing results</b></p>==== | |
| + | <b>Investigator: Achim, Anna <br /></b> | ||
| + | <p style="font-size:13px; color:#66bbff;">Comment: Sequencing results of Loop insertion BioBricks from 31.08.10; the following clones were sent for sequencing: </p> | ||
| + | <br /> | ||
| + | *2: 453 BAP: No Insert | ||
| + | [[File:Freiburg10 453 bap.JPG|400px]] | ||
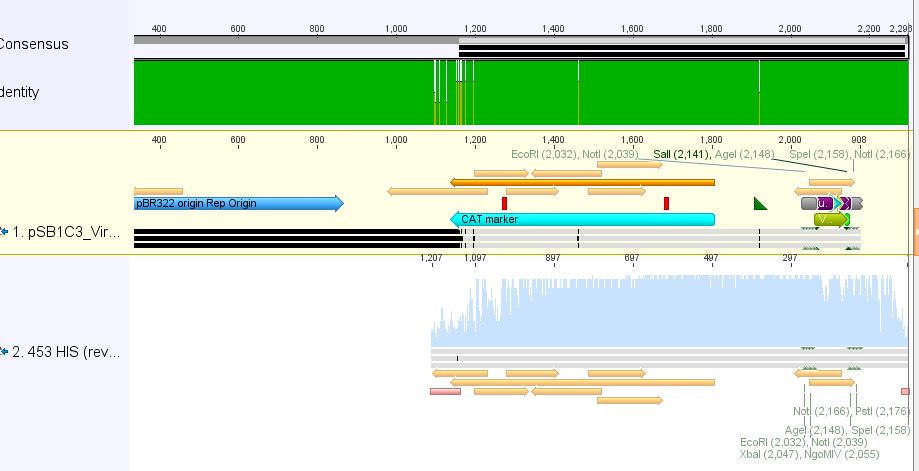
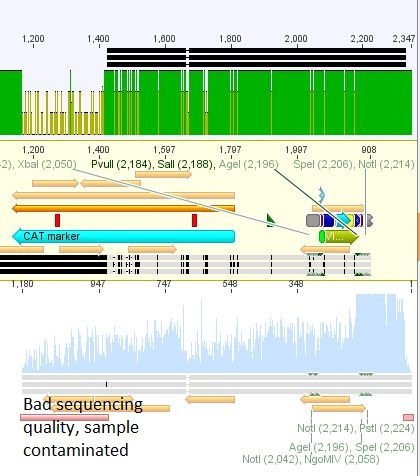
| + | *3:453 His: Insert okay | ||
| + | [[File:Freiburg10 453 his.JPG|400px]] | ||
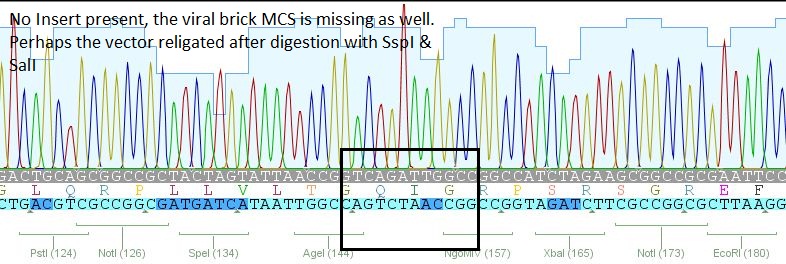
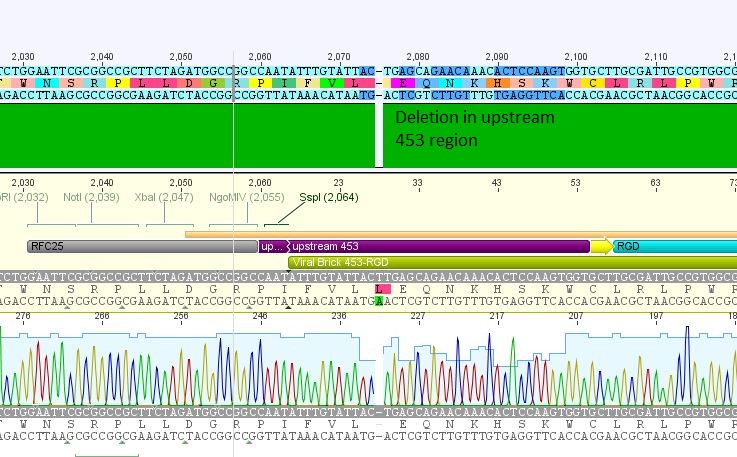
| + | *5: 453 RGD: Mutation in 453 upstream region | ||
| + | [[File:Freiburg10 453 rgd.JPG|400px]] | ||
| + | *7: 587 BAP: Mutation in Bap insert | ||
| + | [[File:Freiburg10 587 bap.JPG|400px]] | ||
| + | *9:587 His: Insert okay | ||
| + | [[File:Freiburg10 587 his.JPG|400px]] | ||
| + | *11:587 KO BAP : Insert okay | ||
| + | [[File:Freiburg10 587 KO bap.JPG|400px]] | ||
| + | *13:587 KO empty : Insert okay | ||
| + | [[File:Freiburg10 587 KO empty.JPG|400px]] | ||
| + | *16: 587 KO his: Mutation in 587 KO | ||
| + | [[File:Freiburg10 587 ko his.JPG|400px]] | ||
| + | *17: 587 KO rgd: No insert | ||
| + | [[File:Freiburg10 587 KO rgd.JPG|400px]] | ||
| + | *19 :587 RGD: Insert okay | ||
| + | [[File:Freiburg10 587 rgd.JPG|400px]] | ||
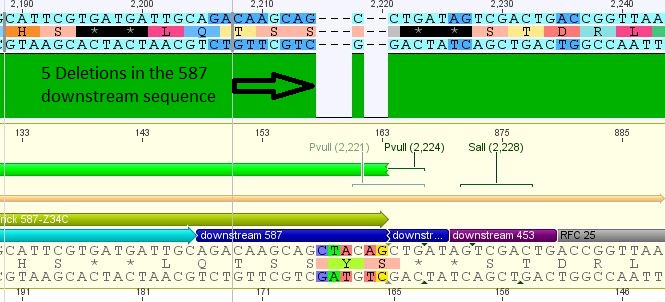
| + | *25: 587 Z34C: Mutation in downstream 587 region | ||
| + | [[File:Freiburg10 587 z34c.JPG|400px]] | ||
| - | + | <b>Mini-Preps:<br /></b> | |
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | New Mini-Preps of the samples with no/mutated insert were prepared. The following clones were used: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | |align="left" | '''Components''' ||align="left"| '''453 BAP''' ||align="left"| '''587 BAP''' ||align="left"| '''453 RGD'''||align="left"| '''587 KO RGD'''||align="left"| '''587 KO HIS'''||align="left"| '''453 Z34C'''||align="left"| '''587 Z34C'''||align="left"| '''587 KO Z34C'''||align="left"| '''587 KO Z34C SPACER''' | |
| - | | | + | |
|- | |- | ||
| - | | align=" | + | | align="left" | Clone||align="left"|1.3 + 1.4 ||align="left"|2.3 + 2.4 ||align="left"|4.3 + 4.4 ||align="left"|6.3 + 6.4 ||align="left"|9.3 + 9.4 ||align="left"|11.3 + 11.4||align="left"|12.2 + 12.4 ||align="left"| 13.2 + 13.4 ||align="left"| 14.3 + 14.4 |
|- | |- | ||
|} | |} | ||
| - | |||
| - | = | + | <br /> |
| + | <p style="font-size:13px; color:#66bbff;"><b> | ||
| + | Comment:</b> Samples 1.3, 2.3, 4.3, 6.3, 9.3, 11.3, 12.2, 13.2, 14.3 were sent for sequencing. For sequencing results go to labday 02.09.10. </p>. | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Design of primers for pAAV_RC</b></p>==== | |
| - | < | + | <b>Investigator: Bea <br /></b> |
| - | + | <p style="font-size:13px; color:#66bbff;">Primers for different cloning strategies of the synthesized cap gene into pAAV_Rc have been designed.</p> | |
| - | + | <ol> | |
| - | + | <li>The first primers are mutagenesis primers which delete the KpnI recognition site at position 3968. These primers can (or should be used) be used if cloning with BspMI/BfuI (isochizomer) does not cut the pAAV_RC efficiently. Instead of using this enzyme we can use XcmI to subclone the cap gene with performing a site-directed mutagenesis afterwards with designed primers. </li> | |
| - | < | + | <li>The second idea is to design oligoduplexes (hybridized oligos) which contain the recognition site for BspMI. In Gormley et al (2002) the idea was to add oligodupexes with the recognition site for BspMI and therefore provide the "second recognition site" in trans. </li> |
| - | + | </ol> | |
| - | + | <b>Next step:</b> Check primers and order them today! Designed primers are stored in my Workingfolder under paav_rc and a word document was created whcih can be found under Oligos --> primers for different strategies... | |
| - | < | + | <br/> |
| - | + | <br/> | |
| - | + | ||
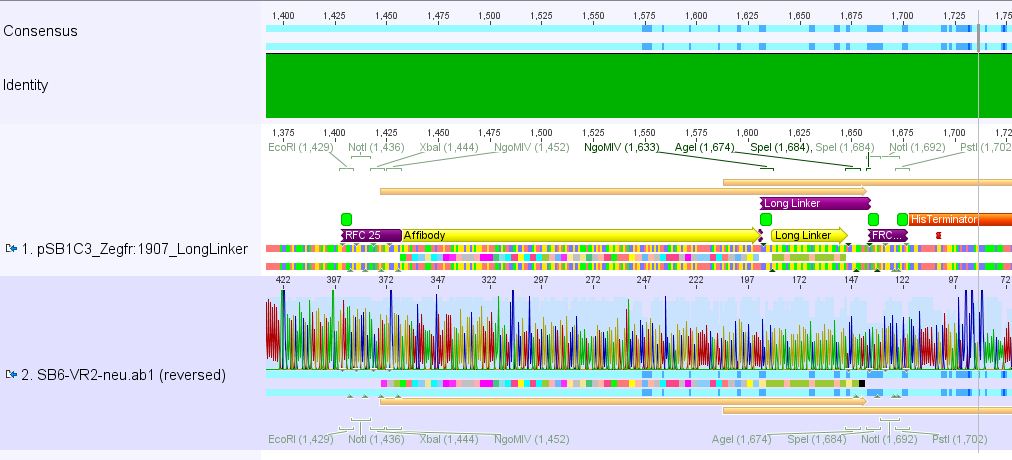
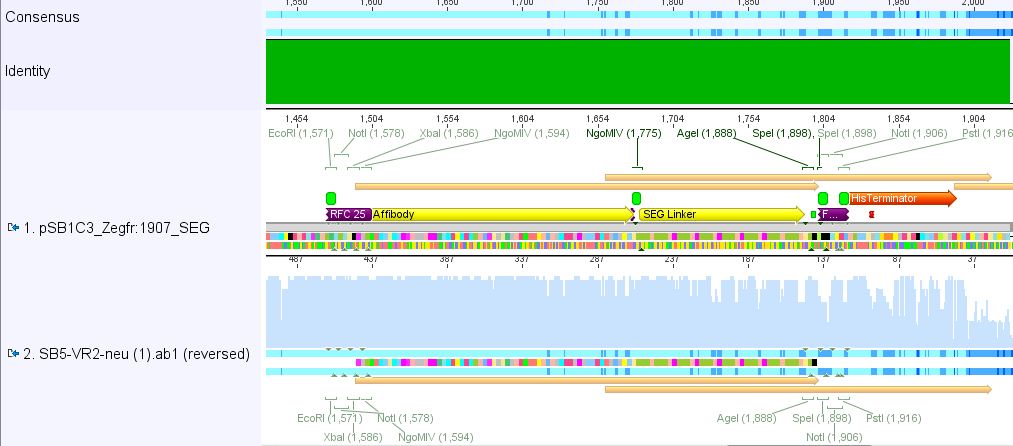
| - | ==== | + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Sequencing results of pSB1C3_Zegfr:1907_Linker</b></p>==== |
| + | <b>Investigator: Hanna<br /></b> | ||
| + | <br/> | ||
| - | + | <p><font color="#00ff00;"><b>Comment:</b> Sequencing results looked all well. One bp was missing in the theoretical sequence of SEG suffix - but is correct in the actual sequence.</font></p> | |
| - | <br> | + | <br/> |
| + | <b><u>pSB1Cr_Zegfr:1907_LongLinker:</u></b> <br/> | ||
| + | [[File:PSB1C3 Zegfr1907 LongLinker.JPG|700px]] | ||
| + | <br/> | ||
| - | + | <b><u>pSB1Cr_Zegfr:1907_SEG:</u></b><br/> | |
| - | + | [[File:PSB1C3 Zegfr1907 SEG.JPG|700px]] | |
| + | <br/> | ||
| - | < | + | <b><u>pSB1Cr_Zegfr:1907_MiddleLinker:</u></b><br/> |
| - | + | [[File:PSB1C3 Zegfr1907 MiddleLinker.JPG|700px]] | |
| - | + | <br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <b><u>pSB1Cr_Zegfr:1907_ShortLinker:</u></b><br/> | |
| - | + | [[File:PSB1C3 Zegfr1907 ShortLinker.JPG|700px]] | |
| + | <br/> | ||
| + | These constructs will be cloned into pCerulean for N-terminal fusion to VP2. | ||
| + | <!--insert this text wherever you want. copy from here until "STOP"--> | ||
| + | <html> | ||
| + | <br /> | ||
| + | <a href="#top">go to the top ;)</a> | ||
| + | </html> | ||
| + | <!--"STOP"--> | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Trafo evalutaion and Inoculation</b></p>==== | ||
| + | <b>Investigator: Kira</b> | ||
| + | <br/> | ||
| + | The plate with transformed CD-biobrick contains around 40 colonies. 4 of them will be inoculated into LB and incubated @37 C over-night. | ||
| + | <!--insert this text wherever you want. copy from here until "STOP"--> | ||
| + | <html> | ||
| + | <br /> | ||
| + | <a href="#top">go to the top ;)</a> | ||
| + | </html> | ||
| + | <!--"STOP"--> | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)</b></p>==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <b>Investigator: Patrick</b> | |
| - | + | ||
| - | + | ||
| - | + | The two Ligations were performed according to the standard protocol:<br> | |
| - | + | 1 µl T4 DNA ligase, 1 µl 10x Buffer, 4,6 µl vector (P273), 3,34 µl Insert (P276, upper and lower band).<br> | |
| - | + | Incubation time: 50 minutes. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | During the transformation i forgot to put the cells into the 37°C shaker so i incubated them afterwards again for 45 minutes at 37 °C on a shaker. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The mutual conles will be picked tomorrow to inoculate 10 ml DYT following a mini-prep on 03.09.2010 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==== | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Transfection, Seeding AAV293 and HT1080</b></p>==== |
| + | <b> Investigator: Kerstin </b> | ||
| - | + | *Transfection: 10 plates, same treatment (10µg of RC (P357), pHelper (P356)and mVenus (P263), following the standard protocol) | |
| - | + | *Seeding HT1080 for transduction with virus from 21.08.2010 (10µg of RC, pHelper, TKGMK; pH=7,10) | |
| + | *Seeding AAV293 for transfection with plasmids without HgH or beta-globin (verifying functionality) | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning of Z<sub>EGFR</sub>:1907_Middle-Linker,Z<sub>EGFR</sub>:1907_Short-Linker, Z<sub>EGFR</sub>:1907_SEG-Linker and Z<sub>EGFR</sub>:1907_Long-Linker into pCerulean</b></p>==== | |
| - | + | <b>Investigator: Stefan </b> | |
| + | <p style="font-size:13px; color:red;"><b>Comment</b>:</p> | ||
| + | <br/> | ||
| + | '''Digestion:''' | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{| border="1" | {| border="1" | ||
| - | | | + | | components || align="right" |Vector (P273) || align="right" |Z<sub>EGFR</sub>:1907_Middle-Linker (P290) || align="right" |Z<sub>EGFR</sub>:1907_Short-Linker (P292) || align="right" |Z<sub>EGFR</sub>:1907_SEG-Linker (P296) || align="right" |Z<sub>EGFR</sub>:1907_Long-Linker (P298) |
|- | |- | ||
| - | | | + | | DNA || align="right" |3,8 || align="right" | 8,8 || align="right" | 10,2 || align="right" | 9,2 || align="right" | 8,7 |
|- | |- | ||
| - | | | + | | BSA (10x) || align="right" |2|| align="right" | 2|| align="right" | 2|| align="right" | 2|| align="right" | 2 |
|- | |- | ||
| - | | | + | | Buffer 4 (10x)|| align="right" |2 || align="right" | 2 || align="right" | 2|| align="right" | 2|| align="right" | 2 |
|- | |- | ||
| - | | | + | |XbaI || align="right" |1 || align="right" |1|| align="right" |1|| align="right" |1|| align="right" |1 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
|- | |- | ||
| - | | align=" | + | |PstI || align="right" |1|| align="right" |1|| align="right" |1|| align="right" |1|| align="right" |1 |
| + | |- | ||
| + | |H<sub>2</sub>O|| align="right" |10,2 || align="right" | 5,2|| align="right" | 3,8|| align="right" | 4,8|| align="right" | 5,3 | ||
| + | |- | ||
| + | |'''Total volume '''|| align="right" |20|| align="right" | 20|| align="right" | 20|| align="right" | 20|| align="right" | 20 | ||
|} | |} | ||
| + | <br> | ||
| + | |||
| + | 0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 50 minutes | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<br /> | <br /> | ||
| + | <br /> | ||
| + | [[File:Freiburg10 pCerulean affi linker.jpg|500px|]] | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | '''Gelextraction:'''<br /> | ||
| + | |||
| + | The gelextraction was performed according to the standard protocol. DNA concentration of the extracts: | ||
| + | * P273: c= 3,1 ng/µl | ||
| + | * P290: c= 3,2 ng/µl | ||
| + | * P292: c= 1,8 ng/µl | ||
| + | * P296: c= 3,0 ng/µl | ||
| + | * P298: c= 3,2 ng/µl | ||
<br> | <br> | ||
| + | '''T4 Ligation:'''<br /> | ||
| - | = | + | The Ligation was performed as following:<br /> |
| + | <p style="font-size:13px; color:red;"><b>Comment</b>: No sufficent vector concentration, therefore 5 µl of vector was used for every approach. </p> | ||
| + | * Vector Volume: 5 µl | ||
| + | * Insert Volume: 3 µl | ||
| - | |||
<br> | <br> | ||
| - | + | * 1µl T4-Ligase buffer (10x) | |
| - | + | * 8µl (Vector + Insert) mix | |
| - | * | + | * 1µl T4-Ligase |
| - | + | <br> Incubating for 40 minutes. | |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | '''Transformation:'''<br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Trafo was performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ===108. labday 02.09.2010=== | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequencing of loop insertions BioBricks: 2nd round</b></p>==== | ||
| + | <b>Investigator: Achim, Anna <br /></b> | ||
| + | <br /> | ||
| + | We analyzed the new sequencing results and found two correct sequences: | ||
| - | < | + | *587 BAP clone 2.3 |
| - | < | + | *453 RGD clone 4.3 |
| - | < | + | |
| - | <p style="font-size:15px; | + | The other samples didn't contain any inserts or had wrong insertions. Apparently, the vector tends to religate easily. A possible reason for the religation of the incompatible ends is the overnight ligation at 18°C. We sent preps of the remaining 7 ligations for sequencing and will prep new clones or repeat the ligation tomorrow, depending on the new sequences. |
| + | |||
| + | <p style="font-size:13px; color:#66bbff;"><b> | ||
| + | Comment:</b> Clones 1.4, 6.4, 9.4, 11.4, 12.4, 13.4, 14.4 were sent for sequencing. For sequencing results go to labday 03.09.10. </p>. | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-preps of pSB1C3_SV40 and pCerulean_VP1_NLS</b></p>==== | ||
| + | <b>Investigator: Bea (Chris L) <br /></b> | ||
<br /> | <br /> | ||
| - | + | Mini-Prep was performed according to the standard protocol | |
<br /> | <br /> | ||
| - | + | <li>P363 = pSB1C3_SV40 clone 1 = 277,70 ng/µl | |
| - | + | <li>P364 = pSB1C3_SV40 clone 2 = 265,01 ng/µl | |
| - | + | <li>P365 = pCerulean_VP1up_NLS (1) clone 1 = 473,14 ng/µl | |
| - | + | <li>P366 = pCerulean_VP1up_NLS (1) clone 2 = 455,88 ng/µl | |
| - | + | <li>P367 = pCerulean_VP1up_NLS (1) clone 3 = 482,86 ng/µl | |
| - | + | <li>P368 = pCerulean_VP1up_NLS (2) clone 1 = 438,52 ng/µl | |
| - | + | <li>P369 = pCerulean_VP1up_NLS (2) clone 2 = 478,00 ng/µl | |
| - | + | <li>P370 = pCerulean_VP1up_NLS (2) clone 3 = 419,00 ng/µl | |
| - | < | + | <br /> |
| - | < | + | Construct were digested with XbaI and PstI for 45 minutes at 37°C. The loading plan on the 1% agarose gel looks like this: |
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br /> | <br /> | ||
| - | + | _M_ ___ _363_ ___ _364_ _365_ _366_ _367_ _368_ _369_ _370_ ___ | |
| - | <br> | + | <br /> |
| - | <br> | + | <br /> |
| + | <b>Results:</b> | ||
| + | P365 was sent for sequencing because test digestion looks goos. In contrast to the SV40 appraoch. This needs to be repeated tomorrow.<br /> | ||
| - | + | <ul> | |
| - | + | <li>Plasmid: pCerulean_VP1up_NLS</li> | |
| - | < | + | <li>Plasmid number: P365</li> |
| - | <p style="font-size:15px; | + | <li>Tube name: SB1</li> |
| - | <p style=" | + | <li>Primer used: CMV-F</li> |
| - | + | </ul> | |
| - | + | ||
| + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Mini-Prep and test digestion of pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker</b></p>==== | ||
| + | <b>Investigator: Hanna <br /></b> | ||
| + | <br/> | ||
| + | <p style="color:#ff00ff;"><b>Comment:</b> Zegfr:1907 (Affibody) and the His-Tag which was coupled to the middle linker were cloned into pCerulean. 3 clones of each construct were picked yesterday. </p> | ||
| + | <br/> | ||
| + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Plasmid Mini-Prep</p> | ||
| + | *new vector name: pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker | ||
<br /> | <br /> | ||
| - | <u>Glycerol Stocks</u> | + | <b><u>Glycerol Stocks</u></b> <br/> |
| + | <b>1. pCerulean_Zegfr:1907</b> | ||
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3 | + | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3''' |
|- | |- | ||
| - | | align="left" | '''Bacteria strain''' ||align="left"| | + | | align="left" | '''Bacteria strain''' ||align="left"| XL1b ||align="left"| XL1b ||align="left"| XL1b |
|- | |- | ||
| - | | align="left" | '''Plasmidname''' ||align="left"| | + | | align="left" | '''Plasmidname''' ||align="left"| pCerulean_Zegfr:1907 ||align="left"| pCerulean_Zegfr:1907 ||align="left"| pCerulean_Zegfr:1907 |
|- | |- | ||
| - | | align="left" | '''Date''' ||align="left"| | + | | align="left" | '''Date''' ||align="left"| 2.9.10 ||align="left"| 2.9.10 ||align="left"| 2.9.10 |
|- | |- | ||
| - | | align="left" | '''given | + | | align="left" | '''given glycerol-stock no.''' ||align="left"| B275 ||align="left"| B276 ||align="left"| B277 |
| + | |- | ||
| + | | align="left" | '''given plasmid no.''' ||align="left"| P371 ||align="left"| P372 ||align="left"| P373 | ||
|} | |} | ||
<br /> | <br /> | ||
| - | < | + | <b>2. pCerulean_6xHis_MiddleLinker</b> |
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3 | + | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3''' |
|- | |- | ||
| - | | align="left" | ''' | + | | align="left" | '''Bacteria strain''' ||align="left"| XL1b ||align="left"| XL1b ||align="left"| XL1b |
| + | |- | ||
| + | | align="left" | '''Plasmidname''' ||align="left"| pCerulean_6xHis_MiddleLinker ||align="left"| pCerulean_6xHis_MiddleLinker ||align="left"| pCerulean_6xHis_MiddleLinker | ||
| + | |- | ||
| + | | align="left" | '''Date''' ||align="left"| 2.9.10 ||align="left"| 2.9.10 ||align="left"| 2.9.10 | ||
| + | |- | ||
| + | | align="left" | '''given glycerol-stock no.''' ||align="left"| B278 ||align="left"| B279 ||align="left"| B280 | ||
| + | |- | ||
| + | | align="left" | '''given plasmid no.''' ||align="left"| P374 ||align="left"| P375 ||align="left"| P376 | ||
|} | |} | ||
<br /> | <br /> | ||
| - | <p style="font-size:15px; font-weight: bold; color: | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Test digestion</p> |
| - | *buffer used: 4 ; Restriction-enzymes used: Enzyme 1 | + | *buffer used: 4; Restriction-enzymes used: Enzyme 1 PstI-HF ; Enzyme 2 EcoRI-HF |
*Plasmid | *Plasmid | ||
| - | **Given Plasmid-Number: | + | **Given Plasmid-Number: 371; DNA concentration: 381.3 ng/µL; |
| - | **Given Plasmid-Number: | + | **Given Plasmid-Number: 372; DNA concentration: 377.5 ng/µL; |
| - | **Given Plasmid-Number: | + | **Given Plasmid-Number: 373; DNA concentration: 409.7 ng/µL; |
| - | + | **Given Plasmid-Number: 374; DNA concentration: 404.2 ng/µL; | |
| + | **Given Plasmid-Number: 375; DNA concentration: 366.1 ng/µL; | ||
| + | **Given Plasmid-Number: 376; DNA concentration: 375.0 ng/µL; | ||
<br /> | <br /> | ||
| - | '''Comments:''' | + | '''Comments:''':) |
<br /> | <br /> | ||
<br /> | <br /> | ||
| - | + | <b>Pipetting scheme for each test digestion:</b> | |
{| border="1" | {| border="1" | ||
| - | | align="left" | | + | | align="left" | <b>Components</b> ||align="left"| <b>Volume/µL</b> |
|- | |- | ||
| - | | align="left" | DNA | + | | align="left" | DNA ||align="left"| 2.7 |
|- | |- | ||
| - | | align="left" | BSA (10x) ||align="left"| | + | | align="left" | BSA (10x) ||align="left"| 1 |
|- | |- | ||
| - | | align="left" | Buffer no. 4 (10x) ||align="left"| 1 | + | | align="left" | Buffer no. 4 (10x) ||align="left"| 1 |
|- | |- | ||
| - | | align="left" | | + | | align="left" | EcoRI-HF ||align="left"| 0.5 |
|- | |- | ||
| - | | align="left" | | + | | align="left" | PstI-HF ||align="left"| 0.5 |
|- | |- | ||
| - | | align="left" | H<sub>2</sub>O ||align="left"| | + | | align="left" | H<sub>2</sub>O ||align="left"| 4.3 |
|- | |- | ||
| - | | align="left" | | + | | align="left" | <b>Total volume</b> ||align="left"| <b>10</b> |
|} | |} | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | *Incubation: 55 minutes | |
| - | + | ||
| - | *Incubation: | + | |
<br /> | <br /> | ||
| - | <p style="font-size:15px; font-weight: bold; color: | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Agarose-Gel:</p> |
<br /> | <br /> | ||
| - | 0. | + | 0.875 g Agarose, 50 mL TAE (1.75 %), 3 µL EthBr, at 115 Volt, running time: 35 minutes |
<br /> | <br /> | ||
<br /> | <br /> | ||
| Line 672: | Line 432: | ||
!Sample | !Sample | ||
!Sample/µl] | !Sample/µl] | ||
| - | !Loading dye ( | + | !Loading dye (6x)/µl |
| - | !Expected size | + | !Expected size |
| - | + | ||
|-- | |-- | ||
| - | | | + | |pCerulean_Zegfr:1907 clone 1 |
| - | | | + | |10 µl |
| - | | | + | |2 µl |
| - | | | + | |231 bp |
| - | + | ||
| - | + | ||
|-- | |-- | ||
| - | | | + | |pCerulean_Zegfr:1907 clone 2 |
| - | | | + | |10 µl |
| - | | | + | |2 µl |
| - | | | + | |231 bp |
| - | + | ||
| - | + | ||
|-- | |-- | ||
| - | | | + | |pCerulean_Zegfr:1907 clone 3 |
| - | | | + | |10 µl |
| - | | | + | |2 µl |
| - | | | + | |231 bp |
| - | | | + | |-- |
| + | |pCerulean_6xHis_MiddleLinker clone 1 | ||
| + | |10 µl | ||
| + | |2 µl | ||
| + | |105 bp | ||
| + | |-- | ||
| + | |pCerulean_6xHis_MiddleLinker clone 2 | ||
| + | |10 µl | ||
| + | |2 µl | ||
| + | |105 bp | ||
| + | |-- | ||
| + | |pCerulean_6xHis_MiddleLinker clone 3 | ||
| + | |10 µl | ||
| + | |2 µl | ||
| + | |105 bp | ||
|-- | |-- | ||
| - | |||
|} | |} | ||
| Line 705: | Line 473: | ||
{| border="1" | {| border="1" | ||
| | | | ||
| - | !Marker | + | !Marker /µL |
| - | !Sample | + | !Sample P371 /µl |
| - | !Sample | + | !Sample P372 /µl |
| - | !Sample | + | !Sample P373 /µl |
| + | !Sample P374 /µl | ||
| + | !Sample P375 /µl | ||
| + | !Sample P376 /µl | ||
|- | |- | ||
!Lane | !Lane | ||
| - | | | + | |5 |
| - | | | + | |12 |
| - | | | + | |12 |
| - | + | |12 | |
| + | |12 | ||
| + | |12 | ||
| + | |12 | ||
|- | |- | ||
|} | |} | ||
<br /> | <br /> | ||
| - | <br> | + | '''Comments:''' Test digestion looked well: <br/> |
| - | < | + | <br/> |
| - | <p style="font-size:15px; | + | [[File:Freiburg10 pCerulean Zegfr1907andpCerulean 6xHis ML.png|700px]] |
| - | + | <br/> | |
| - | + | Clone 1 of each construct were sent for sequencing (P371 and P374 = HW1 and HW2). Used primer: GATC_std_CMV-F. | |
| - | + | <br/> | |
| - | < | + | |
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"> Midi-Prep of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1 & pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1</p>==== | |
| - | + | ||
| - | + | '''Investigators: Chris W. <br> | |
| - | + | <p style="font-size:13px; color:#003399;"> Midi-Preps of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1=P377 and pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1=P378</p> | |
| - | + | The Midi-Preps were performed according to the standard protocol yielding the following concentrations: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
{| border="1" | {| border="1" | ||
| - | | | + | | plasmid-no. || align="right" |P377|| align="right" |P378 |
|- | |- | ||
| - | | | + | | concentration (ng/µl)|| align="right" |325,04 || align="right" |561,15 |
|- | |- | ||
| - | |||
|} | |} | ||
| - | |||
<br> | <br> | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-Prep and test digestion of the CD biobricks</b></p>==== | |
| - | + | <b>Investigator: Kira <br /></b> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p style="font-size:15px; | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:mistake.png|thumb|right|200px]] | |
| - | + | [[File:Cartoons48.jpg|thumb|right|200px|Ohne Worte *lach*]] | |
| - | + | Wh did you digest with eco and ndeI?? is there a spechía reasonn for it?? if the coonstruct was in the rfc standard could digest it with the igem standard enzymes?? | |
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | | components || align="right" | | + | | components || align="right" |sample /µl |
|- | |- | ||
| - | | DNA || align="right" | 2, | + | | DNA || align="right" | 2,5 |
|- | |- | ||
| - | | BSA ( | + | | BSA (100x) || align="right" | 0 |
|- | |- | ||
| - | | Buffer | + | | Buffer ___4_ (10x)|| align="right" | 1,5 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme NdeI (no.Lab:___)|| align="right" | 0,5 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme EcoRI (no.Lab:___)|| align="right" |0,5 |
|- | |- | ||
| - | |H<sub>2</sub>O|| | + | |H<sub>2</sub>O|| align="right" |10 |
|- | |- | ||
| - | |'''Total volume'''|| align="right" | | + | |'''Total volume (e.g. 15,20,25,30 µl)'''|| align="right" | |15 |
|} | |} | ||
| - | <li> Incubation: 1 | + | <li> Incubation: 1 h |
| + | <br /> | ||
| + | <br /> | ||
| - | + | [[File:Freiburg10 test digestion of CD.jpg|500px|]] | |
<br /> | <br /> | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | Sample 1 was sent for sequencing | |
| - | + | <br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ===109. labday 03.09.2010=== | ||
| + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Miniprep and test digestion of pSB1C3_lITR_pTERT_ßglobin_mVenus_hGH_rITR</b></p>==== | ||
| + | <b>Investigator: Achim <br /></b> | ||
| + | <br/> | ||
| + | Comment: The test digestion did not show the expected fragments of 2000 and 2500 bp. the photo was exposed too long to see distinct bands. Digestion will be repeated. | ||
| - | + | [[File:AA1.png|100px]] | |
| - | + | <br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Sequencing results</b></p>==== | |
| - | < | + | <b>Investigator: Hanna <br /></b> |
| - | < | + | <br/> |
| - | < | + | <b>Comment:</b> pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker were cloned and sent for sequencing yesterday. |
| - | < | + | <br/> |
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | </ | + | |
| + | <b>1. pCerulean_Zegfr:1907:</b> Sequencing looked well. | ||
| + | <br/> | ||
| + | [[File:Freiburg10 pCerulean Zegfr1907 seq.JPG|700px]] | ||
| + | <br/> | ||
| + | <b>2. pCerulean_6xHis_MiddleLinker:</b> Sequencing looked also well. | ||
| + | [[File:Freiburg10 pCeruelan 6xHis MiddleLinker seq.JPG|700px]] | ||
| + | <br/> | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Cloning of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker</b></p>==== |
| - | < | + | <b>Investigator: Hanna <br /></b> |
| - | < | + | <br/> |
| - | < | + | |
| - | <p style=" | + | <p style="color:#ff00ff;"><b>Comment:</b> For N-terminal fusion to VP2, the Affibody - fused to different kinds of linkers - will be cloned into the expression plasmid pCerulean. For imaging CFP, which was fused to the middle linker, will be cloned into pCerulean. </p> <br/> |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Practical Cloning:</p> | |
| - | <p style="font-size:15px; font-weight: bold; color: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | *plasmid: | |
| + | **Vector: name: pCerulean number: P273 | ||
| + | **Insert: name: pSB1C3_Zegfr:1907_LongLinker (P298), pSB1C3_Zegfr:1907_MiddleLinker (P290), pSB1C3_Zegfr:1907_SEG (P296), pSB1C3_Zegfr:1907_ShortLinker (P292), pSB1C3_CFP_MiddleLinker (P276) | ||
| + | *new vector name: pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker | ||
| + | *buffer used: 4 ; Restriction-enzymes used: Enzyme 1 EcoRI-HF ; Enzyme 2 PstI-HF | ||
| + | *DNA concentration (vector): 419.5 ng/µL ; DNA concentration (insert): P298 229.7 ng/µL, P290 227.4 ng/µl; P296 218.7 ng/µL; P292 196.6 ng/µL; P276 207.7 ng/µL | ||
<br /> | <br /> | ||
| - | + | '''Comments:''' No. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
| + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Digestion</p> | ||
{| border="1" | {| border="1" | ||
| - | | | + | | components || align="right" |volume of pCerulean /µl || align="right" |volume of Zegfr:1907_MiddleLinker /µl || align="right"|volume of CFP_MiddleLinker /µL |
|- | |- | ||
| - | + | | DNA || align="right" | 8.3 || align="right" | 7.6 || align="right" | 4.8 | |
|- | |- | ||
| - | + | | BSA (10x) || align="right" | - || align="right" | - || align="right" | - | |
|- | |- | ||
| - | + | | Buffer 4 (10x)|| align="right" | 2 || align="right" | 2 || align="right" | 2 | |
|- | |- | ||
| - | + | |Enzyme 1 EcoRI-HF|| align="right" | 1 || align="right" | 1 || align="right" | 1 | |
|- | |- | ||
| + | |Enzyme 2 PstI-HF|| align="right" | 1 || align="right" | 1 || align="right" | 1 | ||
|- | |- | ||
| - | + | |H<sub>2</sub>O|| align="right" | 7.7 || align="right" | 8.4 || align="right" | 11.2 | |
|- | |- | ||
| - | + | |'''Total volume'''|| align="right" | 20 || align="right" | 20 || align="right" | 20 | |
|} | |} | ||
| + | *Incubation: 1.5 h | ||
| + | <br /><br /> | ||
| - | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Agarose-Gel:</p> | |
<br /> | <br /> | ||
| - | + | 0.8 g Agarose, 50 TAE (1.5 %), 5 µL EthBr, at 70 Volt, running time: ~ 1 hour | |
| - | + | ||
| - | 0. | + | |
<br /> | <br /> | ||
<br /> | <br /> | ||
| Line 980: | Line 621: | ||
!Sample/µl] | !Sample/µl] | ||
!Loading dye (6x)/µl | !Loading dye (6x)/µl | ||
| - | !Expected size 1 | + | !Expected size 1 |
| - | + | ||
|-- | |-- | ||
| - | | | + | |P273 |
| - | | | + | |20 µl |
| - | | | + | |4 µl |
| - | | | + | |3922 bp |
| - | + | ||
|-- | |-- | ||
| - | | | + | |P298 |
| - | | | + | |20 µl |
| - | | | + | |4 µl |
| - | | | + | |273 bp |
| - | + | ||
|-- | |-- | ||
| - | | | + | |P290 |
| - | | | + | |20 µl |
| - | | | + | |4 µl |
| - | | | + | |261 bp |
| - | | | + | |-- |
| + | |P296 | ||
| + | |20 µl | ||
| + | |4 µl | ||
| + | |345 bp | ||
| + | |-- | ||
| + | |P292 | ||
| + | |20 µl | ||
| + | |4 µl | ||
| + | |249 bp | ||
| + | |-- | ||
| + | |P276 | ||
| + | |20 µl | ||
| + | |4 µl | ||
| + | |801 bp | ||
|-- | |-- | ||
| - | |||
|} | |} | ||
{| align=right | {| align=right | ||
| Line 1,010: | Line 661: | ||
| | | | ||
!Marker | !Marker | ||
| - | !Sample | + | !Sample P273 /µl |
| - | !Sample | + | !Sample P276 /µl |
| - | !Sample | + | !Sample P290 /µl |
| + | !Sample P292 /µl | ||
| + | !Sample P296 /µl | ||
| + | !Sample P298 /µl | ||
|- | |- | ||
!Lane | !Lane | ||
| - | | | + | |5 |
| - | | | + | |24 |
| - | | | + | |24 |
| - | | | + | |24 |
| + | |24 | ||
| + | |24 | ||
| + | |24 | ||
|- | |- | ||
|} | |} | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Gel extraction</p> | |
| - | + | <br /> | |
| - | <p style="font-size: | + | Gel measurement: |
| - | + | <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | | align="left" | | + | | align="left" | '''Sample''' |
| + | | align="left" | '''Volume''' | ||
| + | | align="left" | '''Concentration''' | ||
|- | |- | ||
| - | | align="left" | | + | | align="left" | P273 |
| + | | align="left" | 20 | ||
| + | | align="left" | 115.45 ng/µL | ||
|- | |- | ||
| - | | align="left" | | + | | align="left" | P276 |
| + | | align="left" | 20 | ||
| + | | align="left" | 9.91 ng/µL | ||
|- | |- | ||
| - | | align="left" | | + | | align="left" | P298 |
| + | | align="left" | 20 | ||
| + | | align="left" | 5.07 ng/µL | ||
|- | |- | ||
| - | | align="left" | | + | | align="left" | P296 |
| + | | align="left" | 20 | ||
| + | | align="left" | 9.03 ng/µL | ||
| + | |- | ||
| + | | align="left" | P290 | ||
| + | | align="left" | 20 | ||
| + | | align="left" | 10.31 ng/µL | ||
| + | |- | ||
| + | | align="left" | P292 | ||
| + | | align="left" | 20 | ||
| + | | align="left" | 20.78 ng/µL | ||
|} | |} | ||
| + | <br /><br /> | ||
| + | |||
| + | |||
| + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Ligation</p> | ||
<br /> | <br /> | ||
| - | < | + | <b>1. PCerulean_Zegfr:1907_LongLinker</b> |
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| ''' | + | | align="left" | ||align="left"| '''P298''' ||align="left"| '''P273''' |
|- | |- | ||
| - | | align="left" | ''' | + | | align="left" | '''Volume/µl''' ||align="left"| 6.61 ||align="left"| 1.39 |
|} | |} | ||
<br /> | <br /> | ||
| - | + | <b>2. PCerulean_Zegfr:1907_MiddleLinker</b> | |
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| ''' | + | | align="left" | ||align="left"| '''P290''' ||align="left"| '''P273''' |
|- | |- | ||
| - | | align="left" | ''' | + | | align="left" | '''Volume/µl''' ||align="left"| 5.53 ||align="left"| 2.47 |
| - | | | + | |} |
| - | | align="left" | ''' | + | <br /> |
| - | + | <b>3. PCerulean_Zegfr:1907_SEG</b> | |
| - | | align="left" | ''' | + | {| border="1" |
| + | | align="left" | ||align="left"| '''P296''' ||align="left"| '''P273''' | ||
|- | |- | ||
| - | | align="left" | ''' | + | | align="left" | '''Volume/µl''' ||align="left"| 6.17 ||align="left"| 1.83 |
|} | |} | ||
<br /> | <br /> | ||
| - | < | + | <b>4. PCerulean_Zegfr:1907_ShortLinker</b> |
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| ''' | + | | align="left" | ||align="left"| '''P292''' ||align="left"| '''P273''' |
|- | |- | ||
| - | | align="left" | ''' | + | | align="left" | '''Volume/µl''' ||align="left"| 4.11 ||align="left"| 3.89 |
|} | |} | ||
<br /> | <br /> | ||
| - | < | + | <b>5. PCerulean_CFP_MiddleLinker</b> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | | align="left" | | + | | align="left" | ||align="left"| '''P276''' ||align="left"| '''P273''' |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | + | | align="left" | '''Volume/µl''' ||align="left"| 7.02 ||align="left"| 0.98 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | align="left" | ''' | + | |
|} | |} | ||
<br /> | <br /> | ||
| - | + | <p style="font-size:15px; font-weight: bold; color: #ff00ff;">Trafo</p> | |
<br /> | <br /> | ||
| - | + | Trafo was performed following the standard protocol. Used cells: XL1b, DNA amount: 2.5 µL of each ligation reaction. Plates were stored @ 37°C room over night. | |
| - | + | <br/> | |
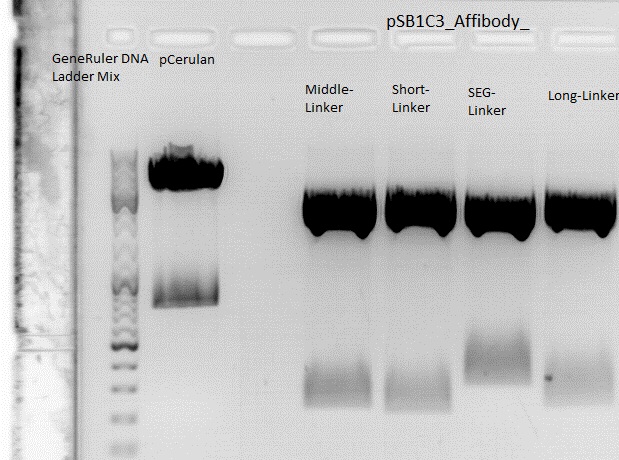
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Harvest viral particles, Seeding HT1080 cells</b></p>==== | ||
| + | <b>Investigator: Adrian, Kerstin <br /></b> | ||
| - | + | *Harvest viral particles of Transfection from 31.08.2010 (six approaches: 10µg of RC (2x P326 and 2x P325 and 2x Stratagene), pHelper and GOI (P262)) | |
| - | + | *Seeding HT1080 cells | |
| - | + | ||
| - | <p style="font-size: | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)</b></p>==== |
| - | + | Investigator: Patrick | |
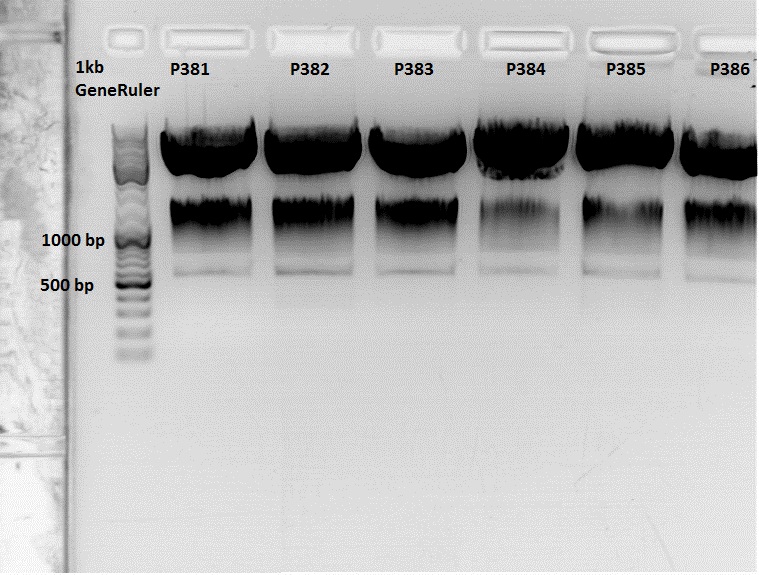
| - | * | + | <br /> |
| - | * | + | The Miniprep was performed according to the standard protocol. |
| - | * | + | Labelling: |
| - | * | + | *P381: pCerulean_middlelinker clone 1 upper band : 526,0 ng/µl |
| - | + | *P382: pCerulean_middlelinker clone 2 upper band : 522,0 ng/µl | |
| - | + | *P383: pCerulean_middlelinker clone 3 upper band : 463,0 ng/µl | |
| - | + | *P384: pCerulean_middlelinker clone 1 lower band : 465,0 ng/µl | |
| - | + | *P385: pCerulean_middlelinker clone 2 lower band : 500,0 ng/µl | |
| - | + | *P386: pCerulean_middlelinker clone 3 lower band : 524,0 ng/µl | |
| + | [[File:Cartoons48.jpg|thumb|right| Öhm Patrick: Hier ist der Grund, warum das Experiment "wiederholt" wurde: Falsche Beschrieftung hier, als auch in der Excel Tabelle ;) "Finde den Fehler!" :P]] | ||
<br> | <br> | ||
| - | + | see also http://www.molbiotech.uni-freiburg.de/iGEM/wiki2010/index.php/August_2010#Cloning_CFP_middlelinker_.28from_pSB1C3_CFP_middlelinker.2C_P276.29_into_pCerulean_.28P273.29 | |
| - | + | [[File:Mistake.png|thumb|right|400px| ;-) bitte ausfüllen]] | |
| - | + | Testdigestion: 7 µl DNA sample, 1 µl Buffer 4 (10x), 1 µl BSA, 0,5 µl Xba, 0,5 µl PstI-HF | |
<br> | <br> | ||
| - | + | Expected size of the fragments: 786 & 2053bp | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | There seems to be something wrong. | |
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | + | [[File:Freiburg10_pat20100903.jpg|400px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Sent for sequencing: P381 & P385 | |
| - | + | Labelling: P381-CMV_rev & P385-CMV_rev <br> | |
| - | + | Used primer: O21 : CMV_reverse_qPCR | |
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning of pSB1C3_leftITR_pTert and pSB1C3_mGMK with pSB1C3_leftITR_CMV</b></p>==== | |
| + | <b>Investigator: Chris L. <br /></b> | ||
| - | + | *Vector: name: pSB1C3_leftITR_pTERT clone 1 '''P256''' c=179,2 ng/µl | |
| - | + | *Vector: name: pSB1C3_leftITR_CMV '''P188''' c=231,6 ng/µl | |
| - | + | *Insert: name: pSB1C3_mGMK '''P195''' c=258,6 ng/µl | |
| + | *new vector name: pSB1C3_leftITR_CMV_GMK | ||
| + | *new vector name: pSB1C3_leftITR_pTERT_GMK | ||
| + | *buffer used: 4 ; Restriction-enzymes used: Enzyme 1 PstI ; Enzyme 2 SpeI ; Enzyme 3 XbaI | ||
| + | <br /> | ||
| + | {| border="1" | ||
| + | | '''components''' || align="right" |'''P195''' || align="right" |'''P256 /µl''' || align="right" |'''P188 /µl''' | ||
| + | |- | ||
| + | | DNA || align="right" | 7,7 || align="right" | 11,2 || align="right" | 8,6 | ||
| + | |- | ||
| + | | BSA (10x) || align="right" | 2 || align="right" | 2 || align="right" | 2 | ||
| + | |- | ||
| + | | Buffer 4 (10x)|| align="right" |2 || align="right" |2 || align="right" |2 | ||
| + | |- | ||
| + | |Enzyme SpeI || align="right" |1 || align="right" |1 || align="right" |0 | ||
| + | |- | ||
| + | |Enzyme XbaI || align="right" |0 || align="right" |0 || align="right" |1 | ||
| + | |- | ||
| + | |Enzyme PstI || align="right" |1 || align="right" |1 || align="right" |1 | ||
| + | |- | ||
| + | |H2O|| align="right" | 6,3 || align="right" | 2,8 || align="right" | 5,4 | ||
| + | |- | ||
| + | |'''Total volume (e.g. 15,20,25,30 µl)'''|| align="right" | 20|| align="right" |20 || align="right" |20 | ||
| + | |} | ||
| - | + | <br /> | |
| - | <br> | + | 0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 130 Volt, running time:45 |
| + | <br /> | ||
<br> | <br> | ||
| + | '''Results''': <br> | ||
| + | [[File:Freiburg10 pSB1C3 lITR pTERT and pSB1C3 lITR CMV with GMK Insert.jpg|550px]] <br> | ||
| + | <br /> | ||
| - | = | + | *c(Insert)= 15,37 ng/µl; size: 627 bp <br /> |
| - | + | *c(Vector)= 50,84 ng/µl; size: 2869 bp | |
| + | *c(Vector)= 75,64 ng/µl; size: 2672 bp | ||
| + | <br /> | ||
| - | + | *Ligation of PCR products and vector: | |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | For the Ligation 1µl T4 buffer (10x) and 1µl T4 ligase were used. Incubation time: 45 min |
| + | <br /> | ||
| + | {| border="1" | ||
| + | | ''' ''' || align="right" |'''pSB1C3_leftITR_CMV + GMK /µl'''|| align="right" |''' pSB1C3_leftITR_pTERT + GMK''' | ||
| + | |- | ||
| + | | Vector || align="right" |5,48 || align="right" | 6,21 | ||
| + | |- | ||
| + | | Insert || align="right" |2,52 || align="right" | 1,79 | ||
| + | |- | ||
| + | |} | ||
| + | <br /> | ||
| - | + | *Transformation: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The transformation was done following the standard protocol using XL1 blue cells.<br /> | |
| - | + | <br /> | |
| - | + | ||
| - | <br> | + | |
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequence analysis of pCerulean_VP1up</b></p>==== |
| - | + | <b>Investigator: Bea<br /></b> | |
| - | + | <p style="font-size:13px; color:#66bbff;">The assembled pCerulean_VP1up which serves as the first construct in the VP1 insertion cloning assembly was sent for sequencing. </p> | |
| - | + | ||
| - | + | ||
| - | = | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p style="font-size: | + | |
| - | + | ||
| - | + | ||
| - | The | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<ul> | <ul> | ||
| - | <li> | + | <li>Plasmid used: P365</li> |
| - | <li> | + | <li>Primer used: CMV-F</li> |
| - | <li> | + | <li>Tube name: SB1</li> |
| - | <li> | + | <li>Folder (Geneious): N-terminal Targeting --> pCerulean_VP1up_NLS</li> |
| - | + | ||
| - | + | ||
</ul> | </ul> | ||
| - | + | <br /> | |
| - | <br> | + | <gallery widths=900px heights=300px> |
| - | + | Image:Freiburg 10 SeqAnalysis of pCerulean VP1up NLS 03.09.2010.jpg | |
| - | < | + | </gallery> |
| - | + | <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br> | + | |
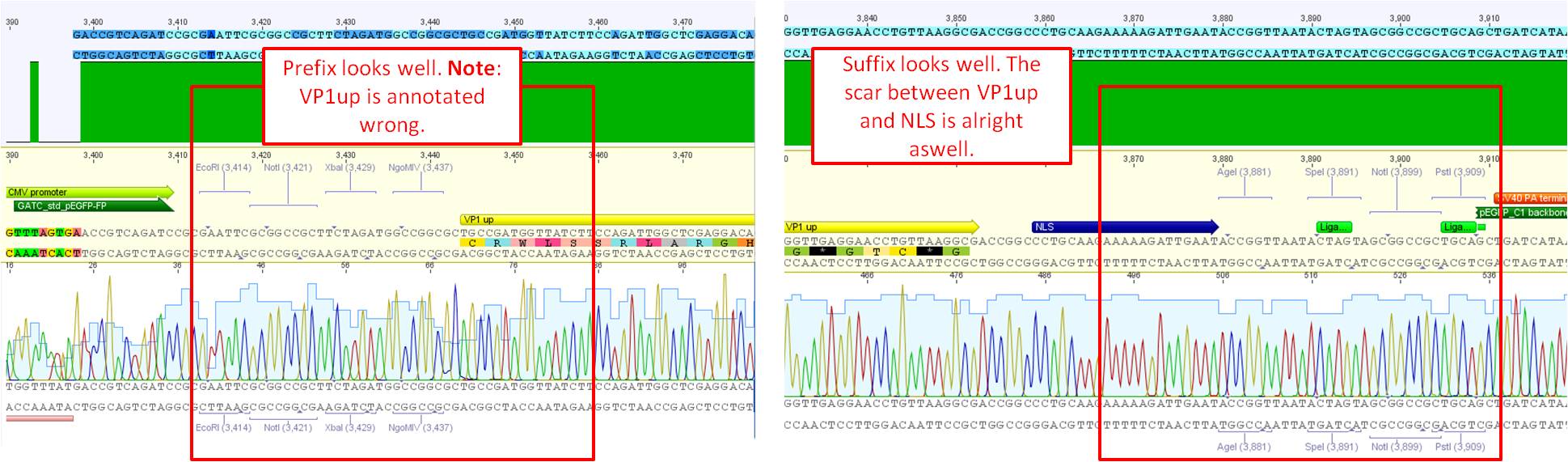
| - | === | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>pCerulean_VP1up_NLS_Targeting molecule</b></p>==== |
| - | + | <b>Investigator: Bea<br /></b> | |
| - | + | <p style="font-size:13px; color:#66bbff;">Comment: For preparing the next step in the VP1 insertion, different targeting molecules will be fused to the construct pCerulean_VP1up_NLS. </p> | |
| - | + | <ol> | |
| - | + | <li>The first construct is the Affibody Z<sub>EGF-R:1907</sub> which was already cloned into the standard iGEM plasmid and can be sued for target tumor cells overexpressing the EGF receptor.</li> | |
| - | + | <li>The second construct is the mVenus (Yellow fluorescent protein) which can be used for imaging experiments</li> | |
| - | + | <li>Another motif will be the His-tag. Since the His-tag is very small, the oligos will be hybridized and directly ligated into the pCerulean backbone.</li> | |
| - | + | </ol> | |
| - | + | <br /> | |
| - | + | <b>Digestion of vector:</b> | |
<ul> | <ul> | ||
| - | <li> | + | <li>Plasmid used: pCerulean_VP1up_NLS (P365) c=470 ng/µL</li> |
| - | <li> | + | <li>Add BSA</li> |
| - | <li> | + | <li>AgeI-HF and SpeI-HF were used</li> |
</ul> | </ul> | ||
| - | + | <br/> | |
| - | + | <b>Protocol of the digestion of the vector:</b> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| border="1" | {| border="1" | ||
| - | + | | align="left" | '''Components''' ||align="left"| <b>v<sub>pCerulean_VP1up_NLS</sub></b>/µL | |
|- | |- | ||
| - | | | + | | align="left" | DNA ||align="left"| 3 |
|- | |- | ||
| - | | | + | | align="left" | BSA (100x) ||align="left"| 2,5 |
|- | |- | ||
| - | | | + | | align="left" | Buffer no. 4 (10x) ||align="left"| 2,5 |
|- | |- | ||
| - | | | + | | align="left" | Enzyme 1 AgeI ||align="left"| 1 |
|- | |- | ||
| - | | | + | | align="left" | Enzyme 2 SpeI HF ||align="left"| 1 |
|- | |- | ||
| - | |H<sub>2</sub>O|| | + | | align="left" | H<sub>2</sub>O ||align="left"| 15 |
|- | |- | ||
| - | |'''Total volume | + | | align="left" | '''Total volume''' ||align="left"| <b>25</b> |
|} | |} | ||
| + | <br /> | ||
| + | <ul> | ||
| + | <li>Incubation of the sample at 37°C</li> | ||
| + | <li>Incubation for 120 minutes</li> | ||
| + | </ul> | ||
| + | <br /> | ||
| + | The Targeting molecules were digested with different enzmyes than the vector for providing the ability to assemble the different constructs together without creating new restriction site but to delete them. This works by fusing NgoMIv and AgeI together. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <b>Digestion of the targeting molecules </b> | ||
| + | <ul> | ||
| + | <li>Plasmids used:</li> | ||
| + | <ol> | ||
| + | <li>pSB1C3_zEGF-R:1907 (P284) c=146 ng/µL</li> | ||
| + | <li>pGA14_mVenus(P60) c=280 ng/µL</li> | ||
| + | </ol> | ||
| + | <li>Add BSA</li> | ||
| + | <li>NgoMIV and SpeI-HF were used</li> | ||
| + | </ul> | ||
| + | <br/> | ||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>v<sub>ZEGFR (P284)</sub></b>/µL ||align="left"| <b>v<sub>pGA_mVenus (P60)</sub></b>/µL | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 13 µl||align="left"| 7 µl | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 2,5 µl||align="left"| 2,5 µl | ||
| + | |- | ||
| + | | align="left" | Buffer no. 4 (10x) ||align="left"| 2,5 µl||align="left"| 2,5 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 1 NgoMIV- ||align="left"| 1,0 µl||align="left"| 1,0 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 2 AgeI HF ||align="left"| 1,0 µl||align="left"| 1,0 µl | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 15 µl||align="left"| 15 µl | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>25</b> ||align="left"| <b>25</b> | ||
| + | |} | ||
| + | <ul> | ||
| + | <li>Incubation of the sample at 37°C</li> | ||
| + | <li>Incubation for 90 minutes</li> | ||
| + | </ul> | ||
| + | <br /> | ||
| + | After incubation the samples were loaded on a 1% agarose gel. The obtained gel (after running the gel for 15 minutes) can be seen below. | ||
| + | <br /> | ||
| + | <gallery widths=400px heights=400px> | ||
| + | Image: Freiburg10 pCerulean VP1up NLS TargetingMolecule.jpg | ||
| + | </gallery> | ||
| + | <br /> | ||
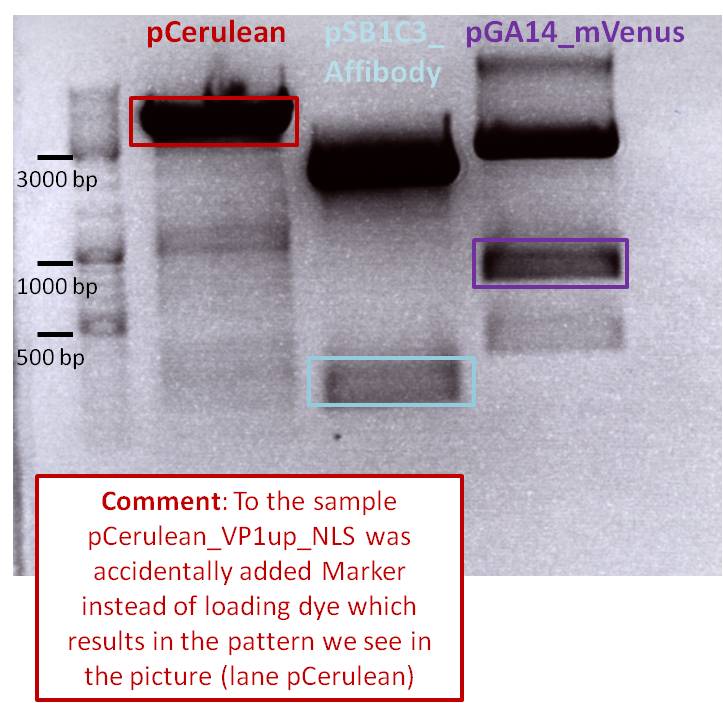
| + | <b>Results: </b>The boxes mark fragments which ere cut out of the gel. The left lane (pCerulean) looks a bit strange, because Marker was added after the digestion reaction instead of Loading dye. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <p style="font-size:13px; color:#33bbff;">Parallel, for assembling the His-Tag to the construct pCerulean_VP1up_NLS, oligos for the His-Tag in the RFC25 standard (provided by Gerrit) were hybridized and can directly be ligated in the digested vector. This strategy was used because of the smal size of the His-Tag which cannot be separated easily by the agarose gel. </p> | ||
| + | <br /> | ||
| + | <b>Hybridization of the His-Oligos</b> | ||
| + | <ul> | ||
| + | <li>10µL oligo1 (1:10)</li> | ||
| + | <li>10µL oligo2 (1:10)</li> | ||
| + | <li>4µL 100mM Tris-Cl pH 8 </li> | ||
| + | <li>10µL 5mM MgCl<sub>2</sub></li> | ||
| + | <li>8µL H<sub>2</sub>O</li> | ||
| + | </ul> | ||
| + | The used programm for the hybridization: | ||
| + | <ul> | ||
| + | <li>99°C 7´</li> | ||
| + | <li>99°C 1´</li> | ||
| + | <li>-1°C R=0,3°/s </li> | ||
| + | <li>Goto 2 rep 74</li> | ||
| + | <li>Hold 4°C</li> | ||
| + | </ul> | ||
| + | <br/> | ||
| + | After the gel extraction of a T4 ligation was performed. | ||
| + | <ul> | ||
| + | <p style="font-size:13px; color:#cc3300;">Affibody Z<sub>EGF-R:1907</sub>:</p> | ||
| + | v<sub>Cerulean_VP1up_NLS</sub> = 5,82µL<br /> | ||
| + | v<sub>ZEGF-R:1907</sub> = 2,18µL <br /> | ||
| + | <br /> | ||
| + | <p style="font-size:13px; color:#cc3300;">mVenus Z:</p> | ||
| + | v<sub>Cerulean_VP1up_NLS</sub> = 4,04µL<br /> | ||
| + | v<sub>mVenus</sub> = 3,96 µL<br /> | ||
| + | <br /> | ||
| + | <p style="font-size:13px; color:#cc3300;">6xHisTag:</p> | ||
| + | v<sub>Cerulean_VP1up_NLS</sub> = 0,2µL<br /> | ||
| + | v<sub>6xHisTag</sub> = 7,98µL <br /> | ||
| + | </ul> | ||
| + | <br /> | ||
| + | <b>Next steps:</b> Picking clones of the trafo plates (containing Kanamycin) and perform the Mini-Prep. The obtaining constructs finally will be fused to the VP2/3 protein which results in the final construct for the VP1 targeting approach. | ||
| + | <br/> | ||
| + | <br/> | ||
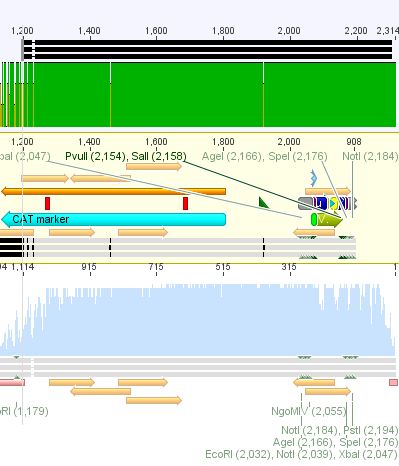
| - | < | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>3rd Repetition of Mini-Preps and test digestion for Loop insertion BioBricks</b></p>==== |
| + | <b>Investigator: Achim, Anna<br /></b> | ||
| - | + | Sequencing results of Loop insertion BioBricks from 02.09.10; the following clones looked well: | |
| - | * | + | *pSB1C3_587_KO_RGD_clone6.4<br> |
| + | *PSB1C3_587_KO_His_clone9.4 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| + | <p style="font-size:13px; color:#003399;"><b>Update</b>: 9 of the 14 ViralBricks looked well (sequencing results from 01. - 03.09.). Two new clones of the remaining constructs were prepared and test digested. </p> | ||
| - | < | + | <b>Test digestion:</b> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| + | {| border="1" | ||
| + | | components ||Volume for each sample /µl | ||
| + | |- | ||
| + | | DNA || 10 | ||
| + | |- | ||
| + | | BSA (10x) ||1,5 | ||
| + | |- | ||
| + | | Buffer 4 (10x) ||1,5 | ||
| + | |- | ||
| + | |Enzyme EcoRI HF ||0,5 | ||
| + | |- | ||
| + | |Enzyme NotI HF ||0,5 | ||
| + | |- | ||
| + | |H2O || 1 | ||
| + | |- | ||
| + | |'''Total volume /µl'''||15 | ||
| + | |} | ||
| + | <br /> | ||
| + | {| border="1" | ||
| - | + | |align="left" | '''Components''' ||align="left"| '''453 BAP''' ||align="left"| '''453 Z34C'''||align="left"| '''587 Z34C'''||align="left"| '''587 KO Z34C'''||align="left"| '''587 KO Z34C SPACER''' | |
| - | + | |- | |
| - | + | | align="left" | Clone||align="left"|1.5 + 1.6 ||align="left"|11.5 + 11.6 ||align="left"|12.5 + 12.6 ||align="left"|13.5 + 13.6||align="left"| 14.5 + 14.6 | |
| - | + | |- | |
| + | |} | ||
| - | < | + | <br/> |
| - | + | Incubation time: 1 h, Incubation temperature: 37° | |
| - | + | <br/> | |
| - | <br> | + | |
| - | + | <b>Preparation of gel:</b><br/> | |
| - | < | + | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes |
| - | + | <br /> | |
| - | + | <br/> | |
| - | File: | + | [[File:3.9.10 AA.png|400px]] |
| - | + | <br /> | |
| - | </ | + | |
| + | <br/> | ||
| + | <b>Expected fragment sizes /bp:</b> <br/> | ||
| + | pSB1C3: 2051 bp<br/> | ||
| + | BAP: ~150<br/> | ||
| + | Z34C: ~ 200<br/> | ||
| + | <br/> | ||
| + | <p style="font-size:13px; color:#003399;"><b>Comment</b>: The following clones were sent for sequencing: 1.6, 12.6 ,13.6 and 14.5.<br/> To do: Retrafo of clone6.4 and clone9.4 and preparation of glycerol stocks. </p> | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning of Cap into pAAV_RC_ins-rep: preparation for SDM</b></p>==== |
| - | + | <b>Investigator: Stefan </b> | |
| - | + | <p style="font-size:13px; color:red;"><b>Comment</b>: Cloning Cap into pAAV still does not work as planned. As another approach Cap can be cut using BsiWI and XcmI and performing a SDM in the part of Cap not cloned into pAAV. Primers are ordered and should arrive on monday to proceed.</p> | |
| - | < | + | <br/> |
| - | + | '''Digestion:''' | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
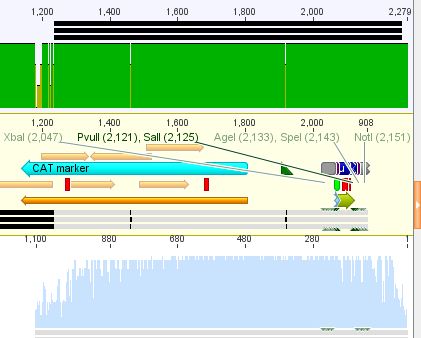
{| border="1" | {| border="1" | ||
| - | | components || align="right" | | + | | components || align="right" |pMA_RepCap Vector_SDM_InsPvuII (P211) || align="right" |pAAV_RC_ins-rep (P250) |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | DNA || align="right" |7,9 || align="right" | 2,1 |
|- | |- | ||
| - | | Buffer | + | | Buffer 2 (10x)|| align="right" |2 || align="right" | 2 |
|- | |- | ||
| - | | | + | |XcmI || align="right" |1 || align="right" |1 |
|- | |- | ||
| - | | | + | |BsiWI || align="right" |1|| align="right" |1 |
|- | |- | ||
| - | |H<sub>2</sub>O|| align="right" | | + | |H<sub>2</sub>O|| align="right" |8,1 || align="right" | 13,9 |
|- | |- | ||
| - | |'''Total volume | + | |'''Total volume '''|| align="right" |20|| align="right" | 20 |
|} | |} | ||
| - | <br> | + | <br /> |
| - | < | + | <p style="font-size:13px; color:red;"><b>Comment</b>:Digestion was performed using two steps, first incubating for 1 hour at 37 °C, afterwards for 1,5 hours at 55 °C.</p><br /> |
| - | < | + | <br /> |
| - | < | + | |
| - | <br> | + | |
| + | '''Gel:''' <br /> | ||
| - | + | 0,5 g Agarose,50 ml TAE (1%), 5 µl EtBr , at 115 Volt, running time: 50 minutes | |
| - | + | <br /> | |
| + | <br /> | ||
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | [[File:Freiburg10 pCerulean pMA pAAV.png|500px|]] | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | <br> | + | |
| - | ''' | + | '''Gelextraction:'''<br /> |
| - | + | ||
| - | + | ||
| - | <br> | + | |
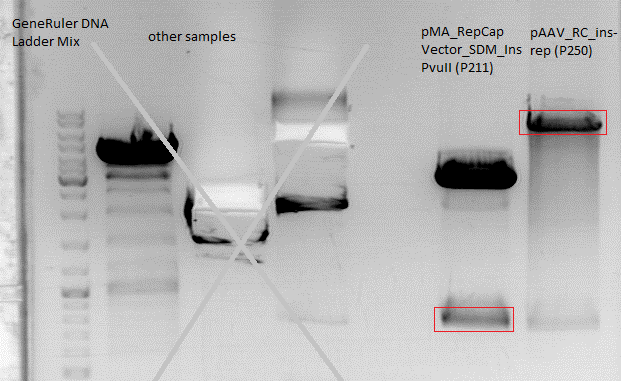
| - | + | The gelextraction was performed according to the standard protocol. DNA concentration of the extracts: | |
| - | + | * pMA_RepCap Vector_SDM_InsPvuII (P211): c= 6,26 ng/µl | |
| + | * pAAV_RC_ins-rep (P250): c= 13,72 ng/µl | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | <p style=" | + | '''Quick Ligation:'''<br /> |
| - | + | <p style="font-size:13px; color:red;"><b>Comment</b>: XcmI produces only 1 base overhang, therefore Quickligase was used for ligation and incubation time increased.</p> | |
| - | The | + | The Ligation was performed as following:<br /> |
| - | + | * Vector Volume: 4,59 µl | |
| - | + | * Insert Volume: 4,41 µl | |
| - | * | + | |
| - | * | + | |
| - | + | ||
<br> | <br> | ||
| - | <br> | + | * 10 µl QuickLigase buffer (2x) |
| + | * 9 µl (Vector + Insert) mix | ||
| + | * 1 µl QuickLigase | ||
| + | <br> Incubating for 60 minutes. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| + | '''Transformation:'''<br /> | ||
| - | + | Trafo was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin. | |
<br> | <br> | ||
| - | |||
| - | |||
| - | + | ===110. labday 04.09.2010=== | |
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273): Sequencing results of P381 and P385</b></p>==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | Investigator:Patrick | |
| - | + | Sequencing results of P381 & P385 Labelling: P381-CMV_rev & P385-CMV_rev: | |
| - | + | A wrong primer was used so there are no relevant sequence data yet. | |
| + | On sunday i will sent these clones again for sequencing. Primer: GATC_std_CMV-F | ||
| - | + | [[Image:Freiburg10_Kopfkratzen.gif|thumb|400px|Ha ich war schneller als Bea beim "Kopfkratzen-setzen" *lol*]] | |
| - | + | '''Hiermit verspreche ich dass es noch jede Menge Kopfkratzer, Laboraufsichtswichtel und pinke Panter geben wird (Patrick)''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <br><br><br><br> | |
| - | <br> | + | <br><br><br><br> |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Harvest viral particles, Transduction</b></p>==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <b> Investigator:Kerstin, Adrian </b> | ||
| - | + | *Harvest viral paricles from one plate of transfection from 01.09.2010 (10 plates, same treatment (10µg of each plasmid P357, P263, P356)) | |
| - | * | + | *Transduction: |
| - | + | ||
| - | + | # 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P326) | |
| - | + | # 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P325) | |
| - | + | # 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 01.09.2010 (stratagene R/C) | |
| - | + | # 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 04.09.2010 (10 plates same treatment) | |
| - | + | # 6x 6-well: 0,5ml of virus from 21.08.2010 (TKGMK; 18x pH 7,10 and 6x pH 7,12) | |
| - | + | ||
| - | + | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Repetition of Hybridisation and Cloning for Loop insertion BioBricks</b></p>==== |
| - | + | ||
| - | + | <b>Investigator: Achim, Anna<br /></b> | |
| - | * | + | *Samples: |
| - | + | 1: 453_BAP | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| + | 11: 453_Z34C<br> | ||
| + | 12: 587_Z34C<br> | ||
| + | 13: 587_KO_Z34C<br> | ||
| + | 14: 587_KO_Z34C_Spacer<br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| + | <p style="font-size:13px; color:#003399;"><b>Comment</b>: New approaches of the Hybridisation and Fill-in reactions were done because it didn't work the first time. A possible reason is that the klenow enzyme wasn't inactivated. In addition, the conditions for ligation will be improved. Also the ligation of 453_BAP was done again.</p> | ||
| - | + | <b>Digestion:</b> | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{| border="1" | {| border="1" | ||
| - | | | + | | components ||Volume Vector 587 ||Volume Vector 453 || Sample 11 || Sample 12 || Sample 13 || Sample 14 |
|- | |- | ||
| - | | | + | | DNA || 3,7 || 3,7|| 6 || 6 || 6 || 6 |
|- | |- | ||
| - | | | + | | BSA (10x) ||-|| -|| -|| -|| -|| - |
| - | + | |- | |
| - | + | | Buffer 4 (10x) ||2|| 2 || 2|| 2|| 2|| 2 | |
| - | + | |- | |
| - | + | |Enzyme 1° ||Bam || Ssp || Ssp || Bam || Bam || Bam | |
| - | + | ||
| - | | | + | |
|- | |- | ||
| - | | | + | |Enzyme 2° ||Pvu|| Sal|| Sal || Pvu || Pvu || Pvu |
| + | |||
|- | |- | ||
| + | |H2O || 12,3 || 12,3 || 10 || 10 || 10 || 10 | ||
| + | |- | ||
| + | |'''Total volume /µl'''||20 | ||
|} | |} | ||
| - | <br> | + | <br /> |
| - | + | ° 1 µl each | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <b>Preparation of gel:</b><br/> |
| - | + | 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 50 minutes | |
| - | === | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Repetition: Prepatation for SDM: cloning of Cap into pAAV</b></p>==== |
| + | Investigator: Stefan | ||
| + | [[Image:Freiburg10_Kopfkratzen.gif|thumb|right|Schönen Abend dir noch]] | ||
| - | <p style="font-size: | + | <p style="font-size:13px; color:red;"><b>Comment</b>: On yesterday's plates nothing grew. Because gelex and ligation products were stored in coldroom, new ligation and trafo approaches were performed. </p> |
| - | + | <br/> | |
| - | + | ||
| + | <br /> | ||
| + | '''Ligation approaches:'''<br /> | ||
| + | * ligation with Quick Ligase (as performed yesterday) | ||
| + | * ligation with T4 Ligase | ||
| + | <br /> | ||
| + | <br /> | ||
| + | '''Transformation approaches:'''<br /> | ||
| + | * Quick Ligase (approach from yesterday): | ||
| + | ** 2 µl of ligation product | ||
| + | ** 4 µl of ligation product<br /> | ||
| + | <br /> | ||
| + | * Quick Ligase (new approach): | ||
| + | ** 2 µl of ligation product | ||
| + | ** 4 µl of ligation product<br /> | ||
| + | <br /> | ||
| + | * T4 Ligase: | ||
| + | ** 2 µl of ligation product | ||
| + | ** 4 µl of ligation product<br /> | ||
| + | <br /> | ||
| + | '''Transformation:'''<br /> | ||
| + | |||
| + | Trafo of each approach was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin. | ||
<br> | <br> | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Ligation and trafo of the modified Cap insert , the Viral Bricks 1, 11, 12, 13 and 14</b></p>==== |
| - | + | ||
| - | + | <b>Investigator: Volker<br /></b> | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p style="font-size:15px; font-weight: bold; color: blue;">Ligation</p> | ||
| + | <br /> | ||
{| border="1" | {| border="1" | ||
| - | | | + | |
| + | |'''Construct''' | ||
| + | |'''Vector (µl)''' | ||
| + | |'''Insert(µl)''' | ||
|- | |- | ||
| - | | | + | |'''ViralBrick 1 in 453''' |
| + | | 7.91 | ||
| + | | 0.09 | ||
|- | |- | ||
| - | | | + | |'''ViralBrick 11 in 453''' |
| - | + | | 6.17 | |
| - | + | | 1.83 | |
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |'''ViralBrick 12 in 587''' |
| + | | 6.15 | ||
| + | | 1.85 | ||
|- | |- | ||
| - | | | + | |'''ViralBrick 13 in 587''' |
| - | + | | 5.3 | |
| - | + | | 2.7 | |
| - | + | ||
| - | + | ||
| - | 2. | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |'''ViralBrick 14 in 587''' |
| + | | 4.46 | ||
| + | | 3.54 | ||
|- | |- | ||
| - | | | + | |'''p211 in pAAV-RC''' |
| - | + | | 5.52 | |
| - | + | | 3.54 | |
| - | + | ||
| - | + | ||
| - | | | + | |
|- | |- | ||
| - | | | + | |'''p211 in pAAV-RC''' |
| + | | 5.52 | ||
| + | | 3.54 | ||
|- | |- | ||
|} | |} | ||
| + | <br /> | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Preparation of PBS Buffer</b></p>==== |
| - | + | Investigator: Volker | |
| + | Two liters of PBS buffer were prepared for the column purification of the viral particles. The following protocol was used: | ||
| + | *dissolve the following in 800ml ddH<sub>2</sub>O: | ||
| + | **8g of NaCl | ||
| + | **0.2g of KCl | ||
| + | **1.44g of Na<sub>2</sub>HP0<sub>$</sub> (1.69 because of the crystal water in the reagent) | ||
| + | **0.24 g of KH<sub>2</sub>PO<sub>4</sub> | ||
| + | *adjust to pH 7.4 | ||
| + | *adjust to 1000ml | ||
| + | *steril filtrate the solution | ||
| - | + | ===111. labday 05.09.2010=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Mini-prep of glycerol stock pMA_RepCap Vector_SDM_InsPvuII clone 1 (B182)</b></p>==== |
| - | + | <b>Investigator: Stefan<br /></b> | |
| - | + | <br /> | |
| - | + | Glycerol stock: Because glycerol stock B182 was not mixed before putting into -80 °C freezer the cells died. Therefore a new glycerol stock was prepared and labled B302. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Mini-Prep was performed according to the standard protocol. Two preps were prepared: | |
| - | + | <br /> | |
| - | + | <li>P363 = pMA_RepCap Vector_SDM_InsPvuII clone 1 (1) = 215,6 ng/µl | |
| - | + | <li>P364 = pMA_RepCap Vector_SDM_InsPvuII clone 1 (2) = 230,5 ng/µl | |
| - | <br> | + | <br /> |
| - | <p style="font-size:15px; | + | ====<p style="font-size:15px; background-color:#ff00ff;"><b>Picking clones of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker</b></p>==== |
| - | Investigator: | + | <b>Investigator: Hanna<br /></b> |
| - | + | <br/> | |
| - | + | 2 clones of each construct (1. pCerulean_Zegfr:1907_ShortLinker, 2. pCerulean_Zegfr:1907_MiddleLinker, 3. pCerulean_Zegfr:1907_LongLinker, 4. pCerulean_Zegfr:1907_SEG and 5. pCerulean_CFP_MiddleLinker) were picked. | |
| - | + | <br/> | |
| - | + | <br/> | |
| - | + | <b>To do tomorrow (6.9.):</b> Mini-Preps and Test digestion (with EcoRI and PstI). | |
| - | + | ===112. labday 06.09.2010=== | |
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Production of Ampicilin and Chloramphenicol</b></p>==== | ||
| + | <b>Investigator: Jessica<br /></b> | ||
| + | <br /> | ||
| + | * 10ml ethanol (70%) containing 1g Amp in 60µl aliquots | ||
| + | * 10ml ethanol (70%) containing 0,25g Cm in 60 µl aliquots | ||
<html> | <html> | ||
</div> | </div> | ||
</html> | </html> | ||
Revision as of 11:32, 6 September 2010

September
107. labday 01.09.2010
Loop insertion BioBricks: Sequencing results
Investigator: Achim, Anna
Comment: Sequencing results of Loop insertion BioBricks from 31.08.10; the following clones were sent for sequencing:
- 2: 453 BAP: No Insert
- 3:453 His: Insert okay
- 5: 453 RGD: Mutation in 453 upstream region
- 7: 587 BAP: Mutation in Bap insert
- 9:587 His: Insert okay
- 11:587 KO BAP : Insert okay
- 13:587 KO empty : Insert okay
- 16: 587 KO his: Mutation in 587 KO
- 17: 587 KO rgd: No insert
- 19 :587 RGD: Insert okay
- 25: 587 Z34C: Mutation in downstream 587 region
Mini-Preps:
New Mini-Preps of the samples with no/mutated insert were prepared. The following clones were used:
| Components | 453 BAP | 587 BAP | 453 RGD | 587 KO RGD | 587 KO HIS | 453 Z34C | 587 Z34C | 587 KO Z34C | 587 KO Z34C SPACER |
| Clone | 1.3 + 1.4 | 2.3 + 2.4 | 4.3 + 4.4 | 6.3 + 6.4 | 9.3 + 9.4 | 11.3 + 11.4 | 12.2 + 12.4 | 13.2 + 13.4 | 14.3 + 14.4 |
Comment: Samples 1.3, 2.3, 4.3, 6.3, 9.3, 11.3, 12.2, 13.2, 14.3 were sent for sequencing. For sequencing results go to labday 02.09.10.
. Design of primers for pAAV_RC
Investigator: Bea
Primers for different cloning strategies of the synthesized cap gene into pAAV_Rc have been designed.
- The first primers are mutagenesis primers which delete the KpnI recognition site at position 3968. These primers can (or should be used) be used if cloning with BspMI/BfuI (isochizomer) does not cut the pAAV_RC efficiently. Instead of using this enzyme we can use XcmI to subclone the cap gene with performing a site-directed mutagenesis afterwards with designed primers.
- The second idea is to design oligoduplexes (hybridized oligos) which contain the recognition site for BspMI. In Gormley et al (2002) the idea was to add oligodupexes with the recognition site for BspMI and therefore provide the "second recognition site" in trans.
Next step: Check primers and order them today! Designed primers are stored in my Workingfolder under paav_rc and a word document was created whcih can be found under Oligos --> primers for different strategies...
Sequencing results of pSB1C3_Zegfr:1907_Linker
Investigator: Hanna
Comment: Sequencing results looked all well. One bp was missing in the theoretical sequence of SEG suffix - but is correct in the actual sequence.
pSB1Cr_Zegfr:1907_MiddleLinker:
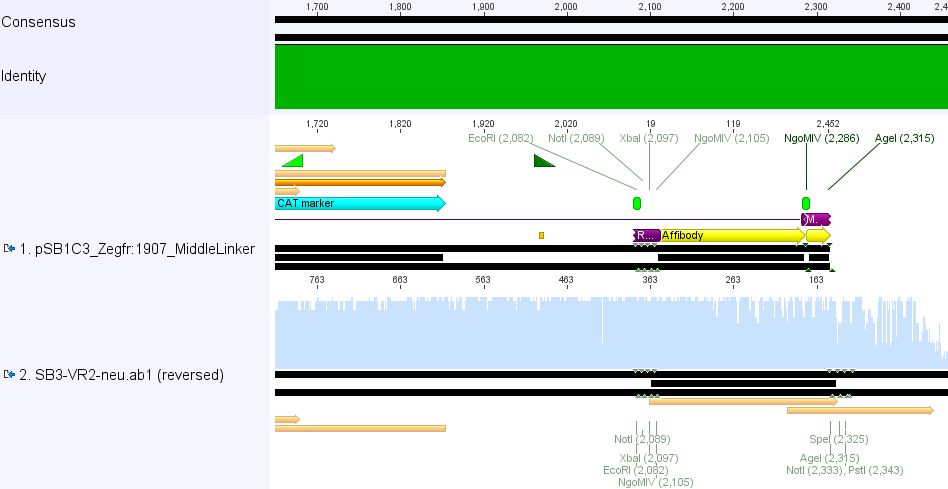
pSB1Cr_Zegfr:1907_ShortLinker:
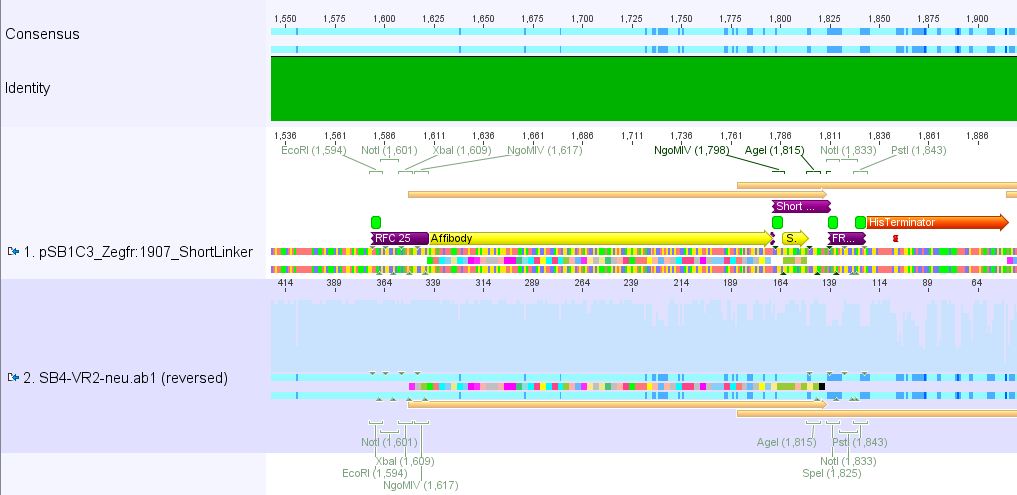
These constructs will be cloned into pCerulean for N-terminal fusion to VP2.
go to the top ;)
Trafo evalutaion and Inoculation
Investigator: Kira
The plate with transformed CD-biobrick contains around 40 colonies. 4 of them will be inoculated into LB and incubated @37 C over-night.
go to the top ;)
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)
Investigator: Patrick
The two Ligations were performed according to the standard protocol:
1 µl T4 DNA ligase, 1 µl 10x Buffer, 4,6 µl vector (P273), 3,34 µl Insert (P276, upper and lower band).
Incubation time: 50 minutes.
During the transformation i forgot to put the cells into the 37°C shaker so i incubated them afterwards again for 45 minutes at 37 °C on a shaker.
The mutual conles will be picked tomorrow to inoculate 10 ml DYT following a mini-prep on 03.09.2010
Transfection, Seeding AAV293 and HT1080
Investigator: Kerstin
- Transfection: 10 plates, same treatment (10µg of RC (P357), pHelper (P356)and mVenus (P263), following the standard protocol)
- Seeding HT1080 for transduction with virus from 21.08.2010 (10µg of RC, pHelper, TKGMK; pH=7,10)
- Seeding AAV293 for transfection with plasmids without HgH or beta-globin (verifying functionality)
Cloning of ZEGFR:1907_Middle-Linker,ZEGFR:1907_Short-Linker, ZEGFR:1907_SEG-Linker and ZEGFR:1907_Long-Linker into pCerulean
Investigator: Stefan
Comment:
Digestion:
| components | Vector (P273) | ZEGFR:1907_Middle-Linker (P290) | ZEGFR:1907_Short-Linker (P292) | ZEGFR:1907_SEG-Linker (P296) | ZEGFR:1907_Long-Linker (P298) |
| DNA | 3,8 | 8,8 | 10,2 | 9,2 | 8,7 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 |
| XbaI | 1 | 1 | 1 | 1 | 1 |
| PstI | 1 | 1 | 1 | 1 | 1 |
| H2O | 10,2 | 5,2 | 3,8 | 4,8 | 5,3 |
| Total volume | 20 | 20 | 20 | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 50 minutes
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P273: c= 3,1 ng/µl
- P290: c= 3,2 ng/µl
- P292: c= 1,8 ng/µl
- P296: c= 3,0 ng/µl
- P298: c= 3,2 ng/µl
T4 Ligation:
The Ligation was performed as following:
Comment: No sufficent vector concentration, therefore 5 µl of vector was used for every approach.
- Vector Volume: 5 µl
- Insert Volume: 3 µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubating for 40 minutes.
Transformation:
Trafo was performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol
108. labday 02.09.2010
Sequencing of loop insertions BioBricks: 2nd round
Investigator: Achim, Anna
We analyzed the new sequencing results and found two correct sequences:
- 587 BAP clone 2.3
- 453 RGD clone 4.3
The other samples didn't contain any inserts or had wrong insertions. Apparently, the vector tends to religate easily. A possible reason for the religation of the incompatible ends is the overnight ligation at 18°C. We sent preps of the remaining 7 ligations for sequencing and will prep new clones or repeat the ligation tomorrow, depending on the new sequences.
Comment: Clones 1.4, 6.4, 9.4, 11.4, 12.4, 13.4, 14.4 were sent for sequencing. For sequencing results go to labday 03.09.10.
. Mini-preps of pSB1C3_SV40 and pCerulean_VP1_NLS
Investigator: Bea (Chris L)
Mini-Prep was performed according to the standard protocol
Construct were digested with XbaI and PstI for 45 minutes at 37°C. The loading plan on the 1% agarose gel looks like this:
_M_ ___ _363_ ___ _364_ _365_ _366_ _367_ _368_ _369_ _370_ ___
Results: P365 was sent for sequencing because test digestion looks goos. In contrast to the SV40 appraoch. This needs to be repeated tomorrow.
- Plasmid: pCerulean_VP1up_NLS
- Plasmid number: P365
- Tube name: SB1
- Primer used: CMV-F
Mini-Prep and test digestion of pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker
Investigator: Hanna
Comment: Zegfr:1907 (Affibody) and the His-Tag which was coupled to the middle linker were cloned into pCerulean. 3 clones of each construct were picked yesterday.
Plasmid Mini-Prep
- new vector name: pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker
Glycerol Stocks
1. pCerulean_Zegfr:1907
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1b | XL1b | XL1b |
| Plasmidname | pCerulean_Zegfr:1907 | pCerulean_Zegfr:1907 | pCerulean_Zegfr:1907 |
| Date | 2.9.10 | 2.9.10 | 2.9.10 |
| given glycerol-stock no. | B275 | B276 | B277 |
| given plasmid no. | P371 | P372 | P373 |
2. pCerulean_6xHis_MiddleLinker
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1b | XL1b | XL1b |
| Plasmidname | pCerulean_6xHis_MiddleLinker | pCerulean_6xHis_MiddleLinker | pCerulean_6xHis_MiddleLinker |
| Date | 2.9.10 | 2.9.10 | 2.9.10 |
| given glycerol-stock no. | B278 | B279 | B280 |
| given plasmid no. | P374 | P375 | P376 |
Test digestion
- buffer used: 4; Restriction-enzymes used: Enzyme 1 PstI-HF ; Enzyme 2 EcoRI-HF
- Plasmid
- Given Plasmid-Number: 371; DNA concentration: 381.3 ng/µL;
- Given Plasmid-Number: 372; DNA concentration: 377.5 ng/µL;
- Given Plasmid-Number: 373; DNA concentration: 409.7 ng/µL;
- Given Plasmid-Number: 374; DNA concentration: 404.2 ng/µL;
- Given Plasmid-Number: 375; DNA concentration: 366.1 ng/µL;
- Given Plasmid-Number: 376; DNA concentration: 375.0 ng/µL;
Comments::)
Pipetting scheme for each test digestion:
| Components | Volume/µL |
| DNA | 2.7 |
| BSA (10x) | 1 |
| Buffer no. 4 (10x) | 1 |
| EcoRI-HF | 0.5 |
| PstI-HF | 0.5 |
| H2O | 4.3 |
| Total volume | 10 |
- Incubation: 55 minutes
Agarose-Gel:
0.875 g Agarose, 50 mL TAE (1.75 %), 3 µL EthBr, at 115 Volt, running time: 35 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size |
|---|---|---|---|
| pCerulean_Zegfr:1907 clone 1 | 10 µl | 2 µl | 231 bp |
| pCerulean_Zegfr:1907 clone 2 | 10 µl | 2 µl | 231 bp |
| pCerulean_Zegfr:1907 clone 3 | 10 µl | 2 µl | 231 bp |
| pCerulean_6xHis_MiddleLinker clone 1 | 10 µl | 2 µl | 105 bp |
| pCerulean_6xHis_MiddleLinker clone 2 | 10 µl | 2 µl | 105 bp |
| pCerulean_6xHis_MiddleLinker clone 3 | 10 µl | 2 µl | 105 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P371 /µl | Sample P372 /µl | Sample P373 /µl | Sample P374 /µl | Sample P375 /µl | Sample P376 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 5 | 12 | 12 | 12 | 12 | 12 | 12 |
Comments: Test digestion looked well:
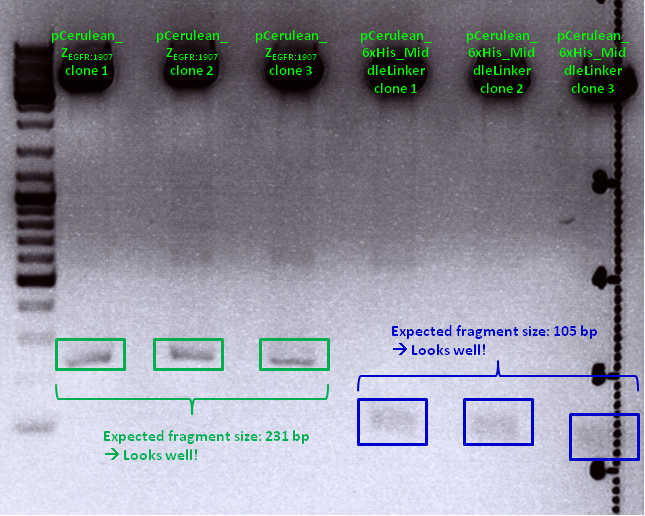
Clone 1 of each construct were sent for sequencing (P371 and P374 = HW1 and HW2). Used primer: GATC_std_CMV-F.
Midi-Prep of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1 & pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1
Investigators: Chris W.
Midi-Preps of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1=P377 and pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1=P378
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P377 | P378 |
| concentration (ng/µl) | 325,04 | 561,15 |
Mini-Prep and test digestion of the CD biobricks
Investigator: Kira
Wh did you digest with eco and ndeI?? is there a spechía reasonn for it?? if the coonstruct was in the rfc standard could digest it with the igem standard enzymes??
| components | sample /µl |
| DNA | 2,5 |
| BSA (100x) | 0 |
| Buffer ___4_ (10x) | 1,5 |
| Enzyme NdeI (no.Lab:___) | 0,5 |
| Enzyme EcoRI (no.Lab:___) | 0,5 |
| H2O | 10 |
| Total volume (e.g. 15,20,25,30 µl) | |15 |
Sample 1 was sent for sequencing
109. labday 03.09.2010
Miniprep and test digestion of pSB1C3_lITR_pTERT_ßglobin_mVenus_hGH_rITR
Investigator: Achim
Comment: The test digestion did not show the expected fragments of 2000 and 2500 bp. the photo was exposed too long to see distinct bands. Digestion will be repeated.
Sequencing results
Investigator: Hanna
Comment: pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker were cloned and sent for sequencing yesterday.
1. pCerulean_Zegfr:1907: Sequencing looked well.
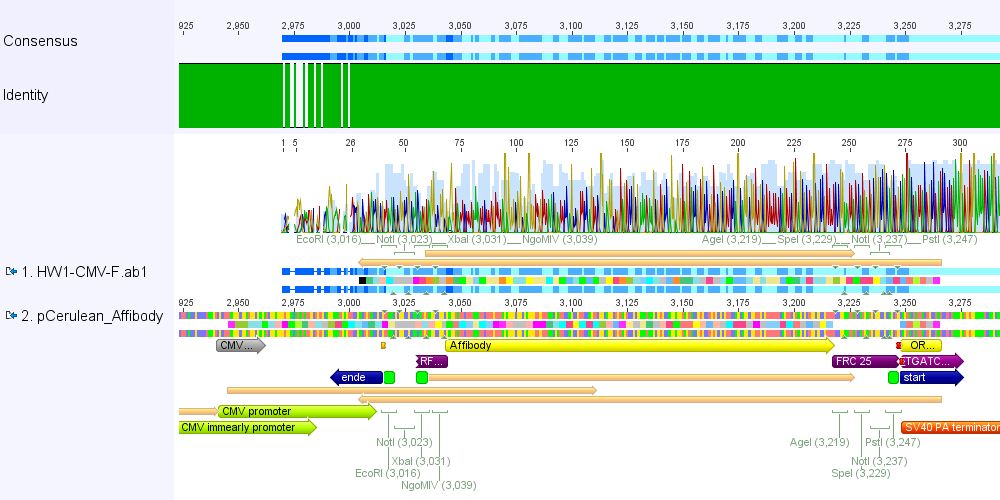
2. pCerulean_6xHis_MiddleLinker: Sequencing looked also well.
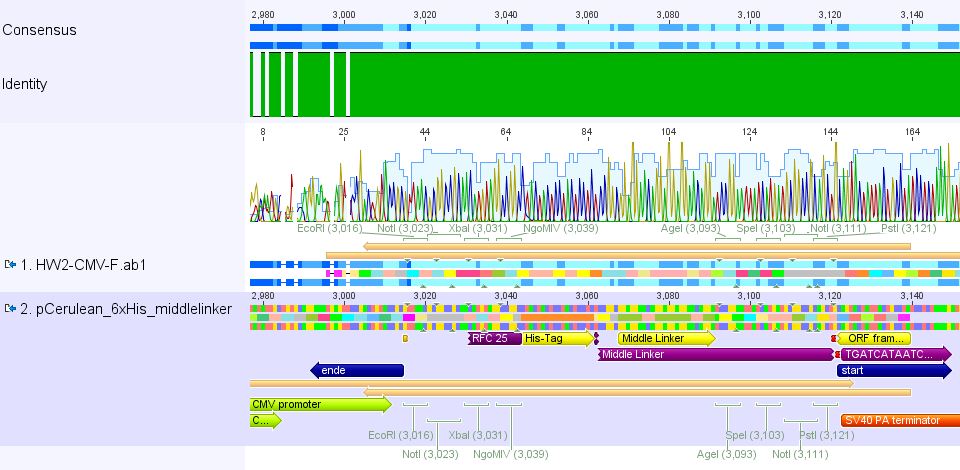
Cloning of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
Investigator: Hanna
Comment: For N-terminal fusion to VP2, the Affibody - fused to different kinds of linkers - will be cloned into the expression plasmid pCerulean. For imaging CFP, which was fused to the middle linker, will be cloned into pCerulean.
Practical Cloning:
- plasmid:
- Vector: name: pCerulean number: P273
- Insert: name: pSB1C3_Zegfr:1907_LongLinker (P298), pSB1C3_Zegfr:1907_MiddleLinker (P290), pSB1C3_Zegfr:1907_SEG (P296), pSB1C3_Zegfr:1907_ShortLinker (P292), pSB1C3_CFP_MiddleLinker (P276)
- new vector name: pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 EcoRI-HF ; Enzyme 2 PstI-HF
- DNA concentration (vector): 419.5 ng/µL ; DNA concentration (insert): P298 229.7 ng/µL, P290 227.4 ng/µl; P296 218.7 ng/µL; P292 196.6 ng/µL; P276 207.7 ng/µL
Comments: No.
Digestion
| components | volume of pCerulean /µl | volume of Zegfr:1907_MiddleLinker /µl | volume of CFP_MiddleLinker /µL |
| DNA | 8.3 | 7.6 | 4.8 |
| BSA (10x) | - | - | - |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme 1 EcoRI-HF | 1 | 1 | 1 |
| Enzyme 2 PstI-HF | 1 | 1 | 1 |
| H2O | 7.7 | 8.4 | 11.2 |
| Total volume | 20 | 20 | 20 |
- Incubation: 1.5 h
Agarose-Gel:
0.8 g Agarose, 50 TAE (1.5 %), 5 µL EthBr, at 70 Volt, running time: ~ 1 hour
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 |
|---|---|---|---|
| P273 | 20 µl | 4 µl | 3922 bp |
| P298 | 20 µl | 4 µl | 273 bp |
| P290 | 20 µl | 4 µl | 261 bp |
| P296 | 20 µl | 4 µl | 345 bp |
| P292 | 20 µl | 4 µl | 249 bp |
| P276 | 20 µl | 4 µl | 801 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample P273 /µl | Sample P276 /µl | Sample P290 /µl | Sample P292 /µl | Sample P296 /µl | Sample P298 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 5 | 24 | 24 | 24 | 24 | 24 | 24 |
Gel extraction
Gel measurement:
| Sample | Volume | Concentration |
| P273 | 20 | 115.45 ng/µL |
| P276 | 20 | 9.91 ng/µL |
| P298 | 20 | 5.07 ng/µL |
| P296 | 20 | 9.03 ng/µL |
| P290 | 20 | 10.31 ng/µL |
| P292 | 20 | 20.78 ng/µL |
Ligation
1. PCerulean_Zegfr:1907_LongLinker
| P298 | P273 | |
| Volume/µl | 6.61 | 1.39 |
2. PCerulean_Zegfr:1907_MiddleLinker
| P290 | P273 | |
| Volume/µl | 5.53 | 2.47 |
3. PCerulean_Zegfr:1907_SEG
| P296 | P273 | |
| Volume/µl | 6.17 | 1.83 |
4. PCerulean_Zegfr:1907_ShortLinker
| P292 | P273 | |
| Volume/µl | 4.11 | 3.89 |
5. PCerulean_CFP_MiddleLinker
| P276 | P273 | |
| Volume/µl | 7.02 | 0.98 |
Trafo
Trafo was performed following the standard protocol. Used cells: XL1b, DNA amount: 2.5 µL of each ligation reaction. Plates were stored @ 37°C room over night.
Harvest viral particles, Seeding HT1080 cells
Investigator: Adrian, Kerstin
- Harvest viral particles of Transfection from 31.08.2010 (six approaches: 10µg of RC (2x P326 and 2x P325 and 2x Stratagene), pHelper and GOI (P262))
- Seeding HT1080 cells
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)
Investigator: Patrick
The Miniprep was performed according to the standard protocol.
Labelling:
- P381: pCerulean_middlelinker clone 1 upper band : 526,0 ng/µl
- P382: pCerulean_middlelinker clone 2 upper band : 522,0 ng/µl
- P383: pCerulean_middlelinker clone 3 upper band : 463,0 ng/µl
- P384: pCerulean_middlelinker clone 1 lower band : 465,0 ng/µl
- P385: pCerulean_middlelinker clone 2 lower band : 500,0 ng/µl
- P386: pCerulean_middlelinker clone 3 lower band : 524,0 ng/µl
Testdigestion: 7 µl DNA sample, 1 µl Buffer 4 (10x), 1 µl BSA, 0,5 µl Xba, 0,5 µl PstI-HF
Expected size of the fragments: 786 & 2053bp
There seems to be something wrong.
Sent for sequencing: P381 & P385
Labelling: P381-CMV_rev & P385-CMV_rev
Used primer: O21 : CMV_reverse_qPCR
Cloning of pSB1C3_leftITR_pTert and pSB1C3_mGMK with pSB1C3_leftITR_CMV
Investigator: Chris L.
- Vector: name: pSB1C3_leftITR_pTERT clone 1 P256 c=179,2 ng/µl
- Vector: name: pSB1C3_leftITR_CMV P188 c=231,6 ng/µl
- Insert: name: pSB1C3_mGMK P195 c=258,6 ng/µl
- new vector name: pSB1C3_leftITR_CMV_GMK
- new vector name: pSB1C3_leftITR_pTERT_GMK
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 PstI ; Enzyme 2 SpeI ; Enzyme 3 XbaI
| components | P195 | P256 /µl | P188 /µl |
| DNA | 7,7 | 11,2 | 8,6 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme SpeI | 1 | 1 | 0 |
| Enzyme XbaI | 0 | 0 | 1 |
| Enzyme PstI | 1 | 1 | 1 |
| H2O | 6,3 | 2,8 | 5,4 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 130 Volt, running time:45
Results:
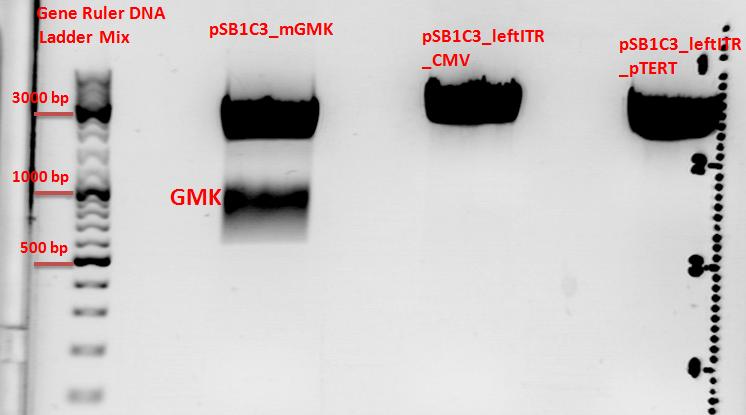
- c(Insert)= 15,37 ng/µl; size: 627 bp
- c(Vector)= 50,84 ng/µl; size: 2869 bp
- c(Vector)= 75,64 ng/µl; size: 2672 bp
- Ligation of PCR products and vector:
For the Ligation 1µl T4 buffer (10x) and 1µl T4 ligase were used. Incubation time: 45 min
| pSB1C3_leftITR_CMV + GMK /µl | pSB1C3_leftITR_pTERT + GMK | |
| Vector | 5,48 | 6,21 |
| Insert | 2,52 | 1,79 |
- Transformation:
The transformation was done following the standard protocol using XL1 blue cells.
Sequence analysis of pCerulean_VP1up
Investigator: Bea
The assembled pCerulean_VP1up which serves as the first construct in the VP1 insertion cloning assembly was sent for sequencing.
- Plasmid used: P365
- Primer used: CMV-F
- Tube name: SB1
- Folder (Geneious): N-terminal Targeting --> pCerulean_VP1up_NLS
pCerulean_VP1up_NLS_Targeting molecule
Investigator: Bea
Comment: For preparing the next step in the VP1 insertion, different targeting molecules will be fused to the construct pCerulean_VP1up_NLS.
- The first construct is the Affibody ZEGF-R:1907 which was already cloned into the standard iGEM plasmid and can be sued for target tumor cells overexpressing the EGF receptor.
- The second construct is the mVenus (Yellow fluorescent protein) which can be used for imaging experiments
- Another motif will be the His-tag. Since the His-tag is very small, the oligos will be hybridized and directly ligated into the pCerulean backbone.
Digestion of vector:
- Plasmid used: pCerulean_VP1up_NLS (P365) c=470 ng/µL
- Add BSA
- AgeI-HF and SpeI-HF were used
Protocol of the digestion of the vector:
| Components | vpCerulean_VP1up_NLS/µL |
| DNA | 3 |
| BSA (100x) | 2,5 |
| Buffer no. 4 (10x) | 2,5 |
| Enzyme 1 AgeI | 1 |
| Enzyme 2 SpeI HF | 1 |
| H2O | 15 |
| Total volume | 25 |
- Incubation of the sample at 37°C
- Incubation for 120 minutes
The Targeting molecules were digested with different enzmyes than the vector for providing the ability to assemble the different constructs together without creating new restriction site but to delete them. This works by fusing NgoMIv and AgeI together.
Digestion of the targeting molecules
- Plasmids used:
- pSB1C3_zEGF-R:1907 (P284) c=146 ng/µL
- pGA14_mVenus(P60) c=280 ng/µL
- Add BSA
- NgoMIV and SpeI-HF were used
| Components | vZEGFR (P284)/µL | vpGA_mVenus (P60)/µL |
| DNA | 13 µl | 7 µl |
| BSA (10x) | 2,5 µl | 2,5 µl |
| Buffer no. 4 (10x) | 2,5 µl | 2,5 µl |
| Enzyme 1 NgoMIV- | 1,0 µl | 1,0 µl |
| Enzyme 2 AgeI HF | 1,0 µl | 1,0 µl |
| H2O | 15 µl | 15 µl |
| Total volume | 25 | 25 |
- Incubation of the sample at 37°C
- Incubation for 90 minutes
After incubation the samples were loaded on a 1% agarose gel. The obtained gel (after running the gel for 15 minutes) can be seen below.
Results: The boxes mark fragments which ere cut out of the gel. The left lane (pCerulean) looks a bit strange, because Marker was added after the digestion reaction instead of Loading dye.
Parallel, for assembling the His-Tag to the construct pCerulean_VP1up_NLS, oligos for the His-Tag in the RFC25 standard (provided by Gerrit) were hybridized and can directly be ligated in the digested vector. This strategy was used because of the smal size of the His-Tag which cannot be separated easily by the agarose gel.
Hybridization of the His-Oligos
- 10µL oligo1 (1:10)
- 10µL oligo2 (1:10)
- 4µL 100mM Tris-Cl pH 8
- 10µL 5mM MgCl2
- 8µL H2O
The used programm for the hybridization:
- 99°C 7´
- 99°C 1´
- -1°C R=0,3°/s
- Goto 2 rep 74
- Hold 4°C
After the gel extraction of a T4 ligation was performed.
Affibody ZEGF-R:1907:
vCerulean_VP1up_NLS = 5,82µL
vZEGF-R:1907 = 2,18µL
mVenus Z:
vCerulean_VP1up_NLS = 4,04µL
vmVenus = 3,96 µL
6xHisTag:
vCerulean_VP1up_NLS = 0,2µL
v6xHisTag = 7,98µL
Next steps: Picking clones of the trafo plates (containing Kanamycin) and perform the Mini-Prep. The obtaining constructs finally will be fused to the VP2/3 protein which results in the final construct for the VP1 targeting approach.
3rd Repetition of Mini-Preps and test digestion for Loop insertion BioBricks
Investigator: Achim, Anna
Sequencing results of Loop insertion BioBricks from 02.09.10; the following clones looked well:
- pSB1C3_587_KO_RGD_clone6.4
- PSB1C3_587_KO_His_clone9.4
Update: 9 of the 14 ViralBricks looked well (sequencing results from 01. - 03.09.). Two new clones of the remaining constructs were prepared and test digested.
Test digestion:
| components | Volume for each sample /µl |
| DNA | 10 |
| BSA (10x) | 1,5 |
| Buffer 4 (10x) | 1,5 |
| Enzyme EcoRI HF | 0,5 |
| Enzyme NotI HF | 0,5 |
| H2O | 1 |
| Total volume /µl | 15 |
| Components | 453 BAP | 453 Z34C | 587 Z34C | 587 KO Z34C | 587 KO Z34C SPACER |
| Clone | 1.5 + 1.6 | 11.5 + 11.6 | 12.5 + 12.6 | 13.5 + 13.6 | 14.5 + 14.6 |
Incubation time: 1 h, Incubation temperature: 37°
Preparation of gel:
1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
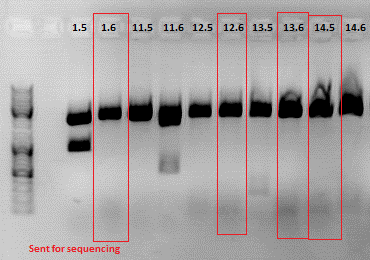
Expected fragment sizes /bp:
pSB1C3: 2051 bp
BAP: ~150
Z34C: ~ 200
Comment: The following clones were sent for sequencing: 1.6, 12.6 ,13.6 and 14.5.
To do: Retrafo of clone6.4 and clone9.4 and preparation of glycerol stocks.
Cloning of Cap into pAAV_RC_ins-rep: preparation for SDM
Investigator: Stefan
Comment: Cloning Cap into pAAV still does not work as planned. As another approach Cap can be cut using BsiWI and XcmI and performing a SDM in the part of Cap not cloned into pAAV. Primers are ordered and should arrive on monday to proceed.
Digestion:
| components | pMA_RepCap Vector_SDM_InsPvuII (P211) | pAAV_RC_ins-rep (P250) |
| DNA | 7,9 | 2,1 |
| Buffer 2 (10x) | 2 | 2 |
| XcmI | 1 | 1 |
| BsiWI | 1 | 1 |
| H2O | 8,1 | 13,9 |
| Total volume | 20 | 20 |
Comment:Digestion was performed using two steps, first incubating for 1 hour at 37 °C, afterwards for 1,5 hours at 55 °C.
Gel:
0,5 g Agarose,50 ml TAE (1%), 5 µl EtBr , at 115 Volt, running time: 50 minutes
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pMA_RepCap Vector_SDM_InsPvuII (P211): c= 6,26 ng/µl
- pAAV_RC_ins-rep (P250): c= 13,72 ng/µl
Quick Ligation:
Comment: XcmI produces only 1 base overhang, therefore Quickligase was used for ligation and incubation time increased.
The Ligation was performed as following:
- Vector Volume: 4,59 µl
- Insert Volume: 4,41 µl
- 10 µl QuickLigase buffer (2x)
- 9 µl (Vector + Insert) mix
- 1 µl QuickLigase
Incubating for 60 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin.
110. labday 04.09.2010
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273): Sequencing results of P381 and P385
Investigator:Patrick
Sequencing results of P381 & P385 Labelling: P381-CMV_rev & P385-CMV_rev: A wrong primer was used so there are no relevant sequence data yet. On sunday i will sent these clones again for sequencing. Primer: GATC_std_CMV-F
Hiermit verspreche ich dass es noch jede Menge Kopfkratzer, Laboraufsichtswichtel und pinke Panter geben wird (Patrick)
Harvest viral particles, Transduction
Investigator:Kerstin, Adrian
- Harvest viral paricles from one plate of transfection from 01.09.2010 (10 plates, same treatment (10µg of each plasmid P357, P263, P356))
- Transduction:
- 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P326)
- 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P325)
- 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 01.09.2010 (stratagene R/C)
- 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 04.09.2010 (10 plates same treatment)
- 6x 6-well: 0,5ml of virus from 21.08.2010 (TKGMK; 18x pH 7,10 and 6x pH 7,12)
Repetition of Hybridisation and Cloning for Loop insertion BioBricks
Investigator: Achim, Anna
- Samples:
1: 453_BAP
11: 453_Z34C
12: 587_Z34C
13: 587_KO_Z34C
14: 587_KO_Z34C_Spacer
Comment: New approaches of the Hybridisation and Fill-in reactions were done because it didn't work the first time. A possible reason is that the klenow enzyme wasn't inactivated. In addition, the conditions for ligation will be improved. Also the ligation of 453_BAP was done again.
Digestion:
| components | Volume Vector 587 | Volume Vector 453 | Sample 11 | Sample 12 | Sample 13 | Sample 14 |
| DNA | 3,7 | 3,7 | 6 | 6 | 6 | 6 |
| BSA (10x) | - | - | - | - | - | - |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1° | Bam | Ssp | Ssp | Bam | Bam | Bam |
| Enzyme 2° | Pvu | Sal | Sal | Pvu | Pvu | Pvu |
| H2O | 12,3 | 12,3 | 10 | 10 | 10 | 10 |
| Total volume /µl | 20 |
° 1 µl each
Preparation of gel:
1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 50 minutes
Repetition: Prepatation for SDM: cloning of Cap into pAAV
Investigator: Stefan
Comment: On yesterday's plates nothing grew. Because gelex and ligation products were stored in coldroom, new ligation and trafo approaches were performed.
Ligation approaches:
- ligation with Quick Ligase (as performed yesterday)
- ligation with T4 Ligase
Transformation approaches:
- Quick Ligase (approach from yesterday):
- 2 µl of ligation product
- 4 µl of ligation product
- Quick Ligase (new approach):
- 2 µl of ligation product
- 4 µl of ligation product
- T4 Ligase:
- 2 µl of ligation product
- 4 µl of ligation product
Transformation:
Trafo of each approach was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin.
Ligation and trafo of the modified Cap insert , the Viral Bricks 1, 11, 12, 13 and 14
Investigator: Volker
Ligation
| Construct | Vector (µl) | Insert(µl) |
| ViralBrick 1 in 453 | 7.91 | 0.09 |
| ViralBrick 11 in 453 | 6.17 | 1.83 |
| ViralBrick 12 in 587 | 6.15 | 1.85 |
| ViralBrick 13 in 587 | 5.3 | 2.7 |
| ViralBrick 14 in 587 | 4.46 | 3.54 |
| p211 in pAAV-RC | 5.52 | 3.54 |
| p211 in pAAV-RC | 5.52 | 3.54 |
Preparation of PBS Buffer
Investigator: Volker
Two liters of PBS buffer were prepared for the column purification of the viral particles. The following protocol was used:
- dissolve the following in 800ml ddH2O:
- 8g of NaCl
- 0.2g of KCl
- 1.44g of Na2HP0$ (1.69 because of the crystal water in the reagent)
- 0.24 g of KH2PO4
- adjust to pH 7.4
- adjust to 1000ml
- steril filtrate the solution
111. labday 05.09.2010
Mini-prep of glycerol stock pMA_RepCap Vector_SDM_InsPvuII clone 1 (B182)
Investigator: Stefan
Glycerol stock: Because glycerol stock B182 was not mixed before putting into -80 °C freezer the cells died. Therefore a new glycerol stock was prepared and labled B302.
Mini-Prep was performed according to the standard protocol. Two preps were prepared:
Picking clones of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
Investigator: Hanna
2 clones of each construct (1. pCerulean_Zegfr:1907_ShortLinker, 2. pCerulean_Zegfr:1907_MiddleLinker, 3. pCerulean_Zegfr:1907_LongLinker, 4. pCerulean_Zegfr:1907_SEG and 5. pCerulean_CFP_MiddleLinker) were picked.
To do tomorrow (6.9.): Mini-Preps and Test digestion (with EcoRI and PstI).
112. labday 06.09.2010
Production of Ampicilin and Chloramphenicol
Investigator: Jessica
- 10ml ethanol (70%) containing 1g Amp in 60µl aliquots
- 10ml ethanol (70%) containing 0,25g Cm in 60 µl aliquots
 "
"