Team:Freiburg Bioware/Project/Project Description
From 2010.igem.org
| Line 62: | Line 62: | ||
| - | <center><h2> | + | <center><h2>Modifications of the Virus Shell via Loop Insertions </h2></center> |
<div style="width:900px; height:400px; border: 1px solid black;"> | <div style="width:900px; height:400px; border: 1px solid black;"> | ||
<div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | <div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | ||
| Line 70: | Line 70: | ||
height="auto"border: 1px solid black;/></div></div> | height="auto"border: 1px solid black;/></div></div> | ||
| - | <center><h2>Specific biotinylation of the Viral | + | <center><h2>Specific biotinylation of the Viral Shell - The Biotinylation Acceptor Peptide (BAP)</h2></center> |
<div style="width:900px; height:400px; border: 1px solid black;"> | <div style="width:900px; height:400px; border: 1px solid black;"> | ||
<div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | <div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | ||
| - | <!---Insert text in here---> | + | <!---Insert text in here---> The BAP (Biotinylation Acceptor Peptide) that we included in our Virus Construction Kit is a 15 amino acid long peptide identified by Schatz J., 1993 in an library screening approach and published under the number #85. This peptide with the sequence 5' - GLNDIFEAQKIEWHE - 3' contains a central lysine that is specifically biotinylated by the prokaryotic enzyme biotin holenzyme synthetase, encoded in the BirA gene of E. coli. Specific biotinylation of this peptide sequence can be performed in vivo by contransfecting a plasmid with the BirA gene as described for the AAV in Arnold et al.; 2006 or by an in vitro coupling approach using the purified Escherichia coli enzyme biotin ligase (BirA). |
| + | The purified BirA biotin ligase that was kindly provided by Avidity. | ||
| + | |||
</p></div> | </p></div> | ||
<div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | <div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | ||
| Line 81: | Line 83: | ||
<div style="width:900px; height:400px; border: 1px solid black;"> | <div style="width:900px; height:400px; border: 1px solid black;"> | ||
<div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | <div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | ||
| - | <!---Insert text in here---> | + | <!---Insert text in here--->Protein tagging via histidine tags is a widely used method for protein purification: Multiple histidine residues (most commonly: Six) are fused to the end of the targeting protein. |
| + | The high binding affinity of histidine towards metal is being exploited for the purification of proteins via the so called “Immobilized Metal Ion Affinity Chromatography“(IMAC): Multiple histidine residues (most commonly: Six) are being fused to the end of the targeting protein. A cell extract containing the recombinant protein is then applied to a column containing immobilized Ni2+-ions. The His-tags complex the Ni2+-ions while other cellular proteins can be washed off the column. The purified proteins can then be eluted with imidazole, which displaces the histidine residues.(M. C. Smith et al. 1988), (Hoffmann & Roeder 1991) | ||
| + | Since the aim behind engineering therapeutic AAV vectors is a safe administration to human patients, it is important to consider a convenient way of purifying the virus particles. Contamination by cellular proteins could cause toxic side effects or a strong immune response. Koerber et al. have first inserted a His-tag into a surface-exposed loop at amino acid position 587 in the Cap protein and successfully purified recombinant virsuses using IMAC (Koerber et al. 2007). For our Virus Construction Kit, we provide the His-tag motif in the ViralBrick standard, allowing for an easy insertion into the 453 and/or 587 loop. | ||
| + | |||
</p></div> | </p></div> | ||
<div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | <div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | ||
| Line 89: | Line 94: | ||
<div style="width:900px; height:400px; border: 1px solid black;"> | <div style="width:900px; height:400px; border: 1px solid black;"> | ||
<div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | <div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | ||
| - | <!---Insert text in here---> | + | <!---Insert text in here---> Integrins are transmembrane proteins that, among other functions, mediate cell attachment to surrounding tissues. They bind to a motif consisting of the amino acids arginine, glycine and aspartic acid (RGD in one-letter code). Because Integrin is highly expressed in many tumor cell lines (S. M. Albelda et al. 1990), (Damjanovich et al. 1992), (Lessey et al. 1995), (Smythe et al. 1995), (Gladson & Cheresh 1991), AAV particles displaying the RGD motif on various positions in their capsid proteins have been created by (Shi et al., 2003). Particles displaying RGD at amino acid positions 584 & 588 as well as 453 or 587 (Boucas et al., 2009) showed transduction efficiencies similar to wt AAV, even when the cells’ HSPG receptors were blocked by heparin sulfate or when the natural HSPG binding motif on the capsid surface was knocked out. To further broaden the area of therapeutic application, we created a ViralBrick containing the RGD motive to specifically target cells with low HSPG-/high Integrin expression. |
</p></div> | </p></div> | ||
<div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | <div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | ||
| Line 98: | Line 103: | ||
<div style="width:900px; height:400px; border: 1px solid black;"> | <div style="width:900px; height:400px; border: 1px solid black;"> | ||
<div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | <div style="float:left; width:460px; height:auto; margin: 0px 5px 0px 5px; text-align:justify;border: 1px solid black;"><p> | ||
| - | <!---Insert text in here---> | + | <!---Insert text in here--->The idea of this targeting approach is to utilize a minimized fragment of the Staphylococcal Protein A that was first described in Staphylococcus aureus. These gram-positive bacteria have evolved the 508 amino acid long protein A that has a high affinity for the Fc-domain of antibodies to protect itself from the immune system. Binding to the constant region of the antibodies is accomplished by the Z-Domain of Protein A that is 58-59 amino acids long, has alone a high affinity (Kd= 14,9 nM) for the antibodies and a three-helix bundle structure. In [Braisted & Wells; 1996] the authors reduced the secundary structure to an two-helix bundle. This size reduction has lead to an drastic reduction of the affinity for IgG (>10^5 fold) which could be recovered by 13 amino acid exchanges resulting in a 38 amino acid long peptide with an satisfying affinity for IgG (Kd = 185 nM) termed Z38. This binding domain was subsequently improved in [Starovasnik et al.; 1997] by the insertion of a disulfide bridge connecting the ends of the helices leading to the binding domain Z34C which shows an increased affinity for IgG (Kd = 20 nM). |
| + | <br> | ||
| + | This engineered antibody binding domain of 34 amino acids was then inserted into capsids of different viral vectors amongst others also the AAV. In [Ried et al.; 2002] the Z34C domain was inserted at position 587 into the capsid of the AAV resulting in viral vector that can be targeted to different target cells without genetic engineering. This targeting approach was then improved in [Gigout et al.; 2005] by the creation of mosaic vectors that contain only ~25% of recombinant VP-Proteins what resulted in 4 to 5 orders of magnitude more infectiosity compared to all-mutant viruses. | ||
</p></div> | </p></div> | ||
<div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | <div style="float:right; width:420px; height:auto; "><img src="www.???.com" width="420" | ||
Revision as of 13:13, 25 October 2010
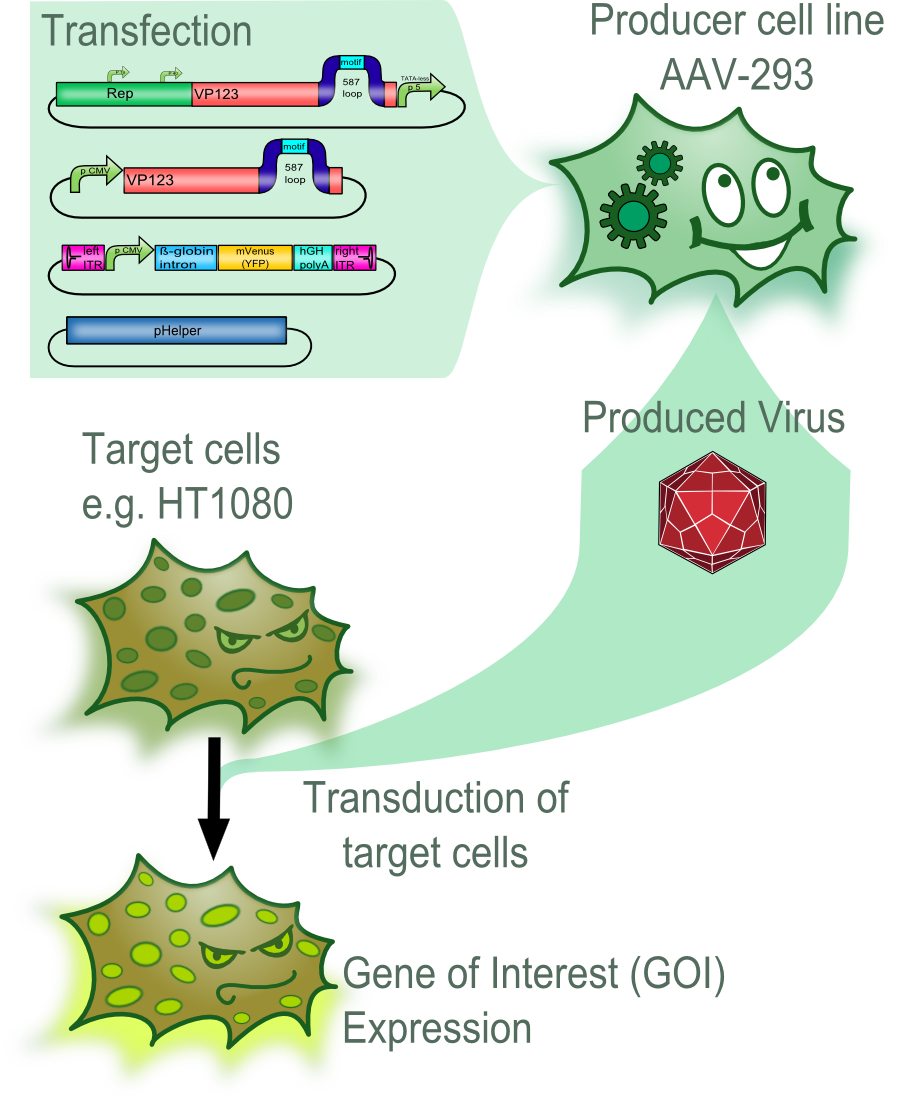
The Experimental System
Therapy using viral vectors is an promising approach for....
In an first step the plasmids of the AAV-2 Helper-free System were genetically modifyed by converting it into BioBricks and inserting of targeting molecules into the constructs. These plasmids were then used to transfect the producer cell line AAV-293. After an incubation of three days the viral vectors were harvested and used to transduce different target cells. The succesful transduction can then for example be measured by detecting the fluorescence of fluorescent proteins in the target cells.
The majority of the modifications that were introduced into the viral vector aimed to allow differential targeting of tumor cell over healthy off-target cells.

Layers of specificity
Employment of viral vectors for means of therapy is idea in the context of personalized medicie that gets more and more interest. In such applications the reduction of side effects and the safety of the patient in general is of the highest priority.
In order to satisfy this requirement we designed our Therapy Vector with several layers of Specificity:
- The targeting of the viral vector towards the desired target cell (e.g. tumor cells) is the basic idea behind the emplyment of viral vectors for therapeutical means. There for the natural tropismn has to be knocked down and a desired tropism has to be introduced that allows differential targeting of pathological but not of off-target cells. To fulfill this mission our Virus Construction Kit offers you different solutions.
- Off-target cells that were transduced by mistake can be preserved from an undesired therapy effect when the therapeutic gene is controley by a tissue specific promoter. For this mean a promoter has to be used that is as specific for the pathological tissue as possible. We included the human telomerase promoter (phTERT) which is often activated in tumor cells and is there for able to allow differential experssion of a therapeutic geneproduct in pathological cells.
- For reasons of safety Therapeutic vector do not directly trigger appoptosis in the successfully targeted cells. To include one further layer of specificity and safety we decided to arm our therapy vector with different prodrug convertases. Neither the single application of the harmless prodrug nor the single expression of the convertase has a noteworthy effect of the transduced cell. Only in cells that express the prodrug convertase and have a sufficient cytoplasmatic concentration of the belonging prodrug apoptosis is triggered. This dependency of the therapy on a prodrug can be employed to protect tissues or other persons that could come in contact with the therapeutical vector. This aspect was specially inportant for the development of a viral vector that is able to infect humans in the context of a undergraduate project for the iGEM competition. Therefor this approach gained our preference over other possibly equivalent arming possibilities described in the tumor therapy with viral vectors.

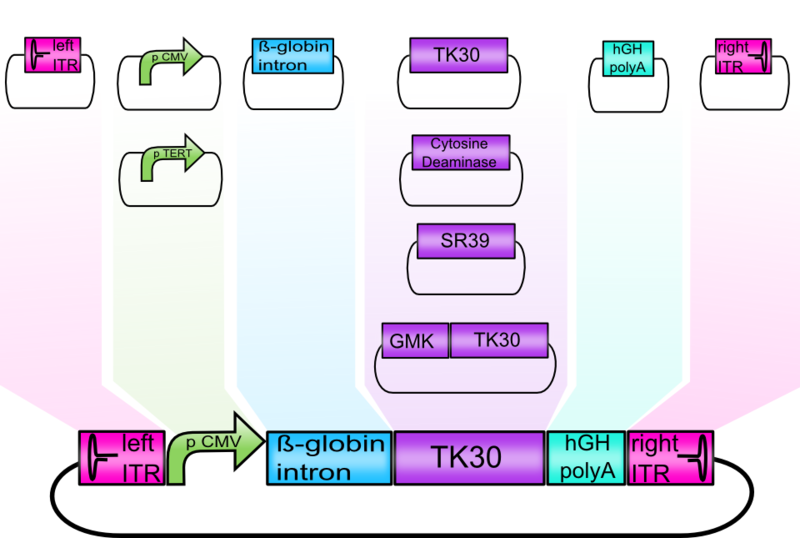
Modularization of the rAAV genome
Text
Modulatization of the RepCap plasmid
Text
Modulatization of the Vectorplasmid
Text

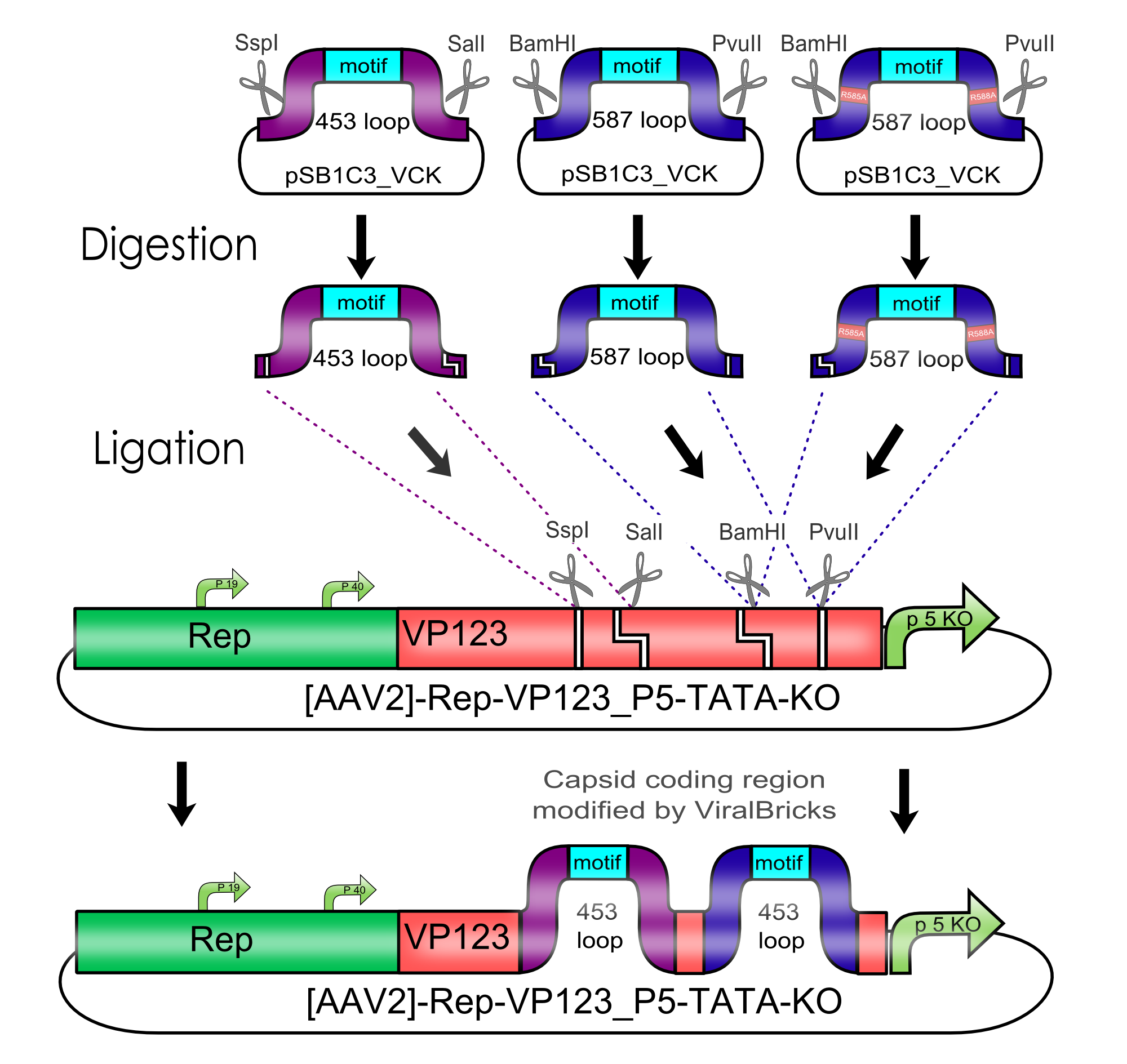
Modification of the viral surface in one cloning step - The ViralBrick standard
AAV bears its natural tropism for HSPG on one of its major exposed surface loops - at amino acid position 585/588. Insertions of small binding motives into this region as well as into another surface loop at amino acid position 453 have proved effective for retargeting the virus towards different receptors.
We introduced two pairs of single cutting restriction sites into each surface loop, allowing an easy swapping of different loop insertions. Our kit comes with a number of those loop insertion motives, labeled ViralBricks. We do not only provide ViralBricks for differential targeting and knocking out of the natural tropism, we also included inserts for purification and detection of virus particles.

Modifications of the Virus Shell via Loop Insertions
Text
Specific biotinylation of the Viral Shell - The Biotinylation Acceptor Peptide (BAP)
The BAP (Biotinylation Acceptor Peptide) that we included in our Virus Construction Kit is a 15 amino acid long peptide identified by Schatz J., 1993 in an library screening approach and published under the number #85. This peptide with the sequence 5' - GLNDIFEAQKIEWHE - 3' contains a central lysine that is specifically biotinylated by the prokaryotic enzyme biotin holenzyme synthetase, encoded in the BirA gene of E. coli. Specific biotinylation of this peptide sequence can be performed in vivo by contransfecting a plasmid with the BirA gene as described for the AAV in Arnold et al.; 2006 or by an in vitro coupling approach using the purified Escherichia coli enzyme biotin ligase (BirA). The purified BirA biotin ligase that was kindly provided by Avidity.
Purification of Therapeutic Viral Vectors - The His-Affinity Tag
Protein tagging via histidine tags is a widely used method for protein purification: Multiple histidine residues (most commonly: Six) are fused to the end of the targeting protein. The high binding affinity of histidine towards metal is being exploited for the purification of proteins via the so called “Immobilized Metal Ion Affinity Chromatography“(IMAC): Multiple histidine residues (most commonly: Six) are being fused to the end of the targeting protein. A cell extract containing the recombinant protein is then applied to a column containing immobilized Ni2+-ions. The His-tags complex the Ni2+-ions while other cellular proteins can be washed off the column. The purified proteins can then be eluted with imidazole, which displaces the histidine residues.(M. C. Smith et al. 1988), (Hoffmann & Roeder 1991) Since the aim behind engineering therapeutic AAV vectors is a safe administration to human patients, it is important to consider a convenient way of purifying the virus particles. Contamination by cellular proteins could cause toxic side effects or a strong immune response. Koerber et al. have first inserted a His-tag into a surface-exposed loop at amino acid position 587 in the Cap protein and successfully purified recombinant virsuses using IMAC (Koerber et al. 2007). For our Virus Construction Kit, we provide the His-tag motif in the ViralBrick standard, allowing for an easy insertion into the 453 and/or 587 loop.
Targeting of integrin overexpressing cells - The RGD Motif
Integrins are transmembrane proteins that, among other functions, mediate cell attachment to surrounding tissues. They bind to a motif consisting of the amino acids arginine, glycine and aspartic acid (RGD in one-letter code). Because Integrin is highly expressed in many tumor cell lines (S. M. Albelda et al. 1990), (Damjanovich et al. 1992), (Lessey et al. 1995), (Smythe et al. 1995), (Gladson & Cheresh 1991), AAV particles displaying the RGD motif on various positions in their capsid proteins have been created by (Shi et al., 2003). Particles displaying RGD at amino acid positions 584 & 588 as well as 453 or 587 (Boucas et al., 2009) showed transduction efficiencies similar to wt AAV, even when the cells’ HSPG receptors were blocked by heparin sulfate or when the natural HSPG binding motif on the capsid surface was knocked out. To further broaden the area of therapeutic application, we created a ViralBrick containing the RGD motive to specifically target cells with low HSPG-/high Integrin expression.
Arming the Viral Vector with therapeutic antibodies - The Z34C Motif
The idea of this targeting approach is to utilize a minimized fragment of the Staphylococcal Protein A that was first described in Staphylococcus aureus. These gram-positive bacteria have evolved the 508 amino acid long protein A that has a high affinity for the Fc-domain of antibodies to protect itself from the immune system. Binding to the constant region of the antibodies is accomplished by the Z-Domain of Protein A that is 58-59 amino acids long, has alone a high affinity (Kd= 14,9 nM) for the antibodies and a three-helix bundle structure. In [Braisted & Wells; 1996] the authors reduced the secundary structure to an two-helix bundle. This size reduction has lead to an drastic reduction of the affinity for IgG (>10^5 fold) which could be recovered by 13 amino acid exchanges resulting in a 38 amino acid long peptide with an satisfying affinity for IgG (Kd = 185 nM) termed Z38. This binding domain was subsequently improved in [Starovasnik et al.; 1997] by the insertion of a disulfide bridge connecting the ends of the helices leading to the binding domain Z34C which shows an increased affinity for IgG (Kd = 20 nM).
This engineered antibody binding domain of 34 amino acids was then inserted into capsids of different viral vectors amongst others also the AAV. In [Ried et al.; 2002] the Z34C domain was inserted at position 587 into the capsid of the AAV resulting in viral vector that can be targeted to different target cells without genetic engineering. This targeting approach was then improved in [Gigout et al.; 2005] by the creation of mosaic vectors that contain only ~25% of recombinant VP-Proteins what resulted in 4 to 5 orders of magnitude more infectiosity compared to all-mutant viruses.
Testing the limit for loop insertion - The Beta-Lactamase
Text
Method Development
Text
Arming: Killing the Tumor
Text
All-In-One: Testing multiple modified Viral Vectors
Text
Virus Construction Kit - The Manual
Text







 "
"