Team:Freiburg Bioware/NoteBook/Labjournal/September2
From 2010.igem.org

Contents |
September
124. labday 19.09.2010
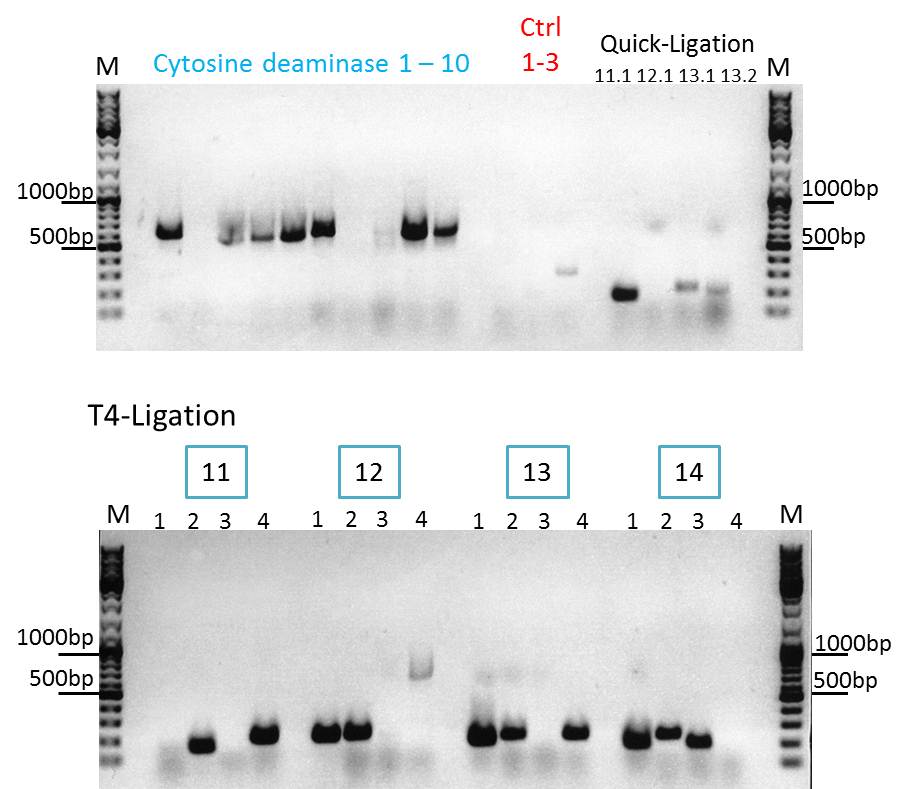
Continuation with Colony-PCR of pSB1C3_CD and ViralBrick motif Z34C
Investigator: Bea
Comment: Since the colony PCR of the last try did not work exactly the way we expected it, another colony PCR approach with new primers was conducted.
Loading plan:
upper lanes
M CD1 CD2 CD3 CD4 CD5 CD6 CD7 CD8 CD9 CD10 +ctrl(CFP) +ctrl(BAP) -ctrl(pAAV_MCS) 11Q1 12Q1 13Q1 13Q2 M
lower lanes
M 11T41 11T42 11T43 11T44 12T41 12T42 12T43 12T44 13T41 13T42 13T43 13T44 14T41 14T42 14T43 14T44 M
Expected sizes of the PCR products are:
- Cytosine deaminase (CD) = 1300bp
- Z34C loops insertion motif = 220bp
- Positive control 1 = CFP = 750bp
- Positive control 2 = BAP_587 = 170bp
Results:
The Colony PCR of the Cytosine Deaminase (CD) did not work out. The detectable bands at around 700 bp correspond to CFP. Therefore, BioBrick production of the CD must be repeated. The following three bands represent the controls. Three different controls were used. The first corresponds to pSB1C3_CFP, the second to BAP_587 and third control is the negative control. Unfortunately now bands can be detected in the postive controls. A faint band can be seen in the negative control which is the only lane in which no band should be detectable.
The following bands after the three controls correspond to the approaches of the Z34C Viralbrick production. Some bands in the lower and the upper lanes show positive results. The corresponding inoculated overnight culture were centrifuged and prepared in order to perform a Mini-Prep tomorrow.
Comment: hmm, seems that the CD-assambly didn't work at all.. Should i try it again or are we gonna skip it for good due to other priorities?(kira)
No, we cannot skip it. Can we change something else?? Any ideas what we can alter in the protocol? Because problem is that we are still waiting for the tk... so we should aswell focus on the other prodrug activating enzmye!! (Bea)
Testtransduction of the final modified capsid coding constructs
Investigator: Adrian
Aim of the experiment:
During the project 22 silent nucleotide exchange mutations were introduced into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. Two point mutations had to be dismissed, because either a first test transduction showed that the construct was not working anymore or the insertion of the synthesized gene posed serious problems, because the restriction enzyme did not work.
Finally, we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carries 20 point mutations. Now we can announce that the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works!
Impression from the moment:
Miniprep of serveral constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B430 = pCerulean_ZEGFR:1907_Longlinker_VP2/3
- B431 = pCerulean_CFP_Middlelinker_VP2/3_insCap
- B432 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap
Mini-Prep was performed according to the standard protocol
- P527 = pCerulean_ZEGFR:1907_Longlinker_VP2/3 c = 230,61 ng/µl
- P528 = pCerulean_CFP_Middlelinker_VP2/3_insCap c = 290,81 ng/µl
- P529 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap c = 269,39 ng/µl
Several other constructs were preped, but need to be confirmed before being added to plasmid/glycerol stock.
125. labday 20.09.2010
Cloning of pCerulean_Affibody_VP2/3 and pSB1C3_Affibody_middlelinker_GFP_his
Investigator: Jessica
Comment:Affibody_VP2/3 will be cloned in pCerulean and Affibody_middlelinker will be cloned together with GFP_His in pSB1C3
- P407= pCerulean_CFP_middlelinker c=482,76 ng/µl
- P516= pSB1C3_ZEGFR:1907_VP2/3 clone 1 c= 191,6 ng/µl
- P290= pSB1C3_Affibody_Middlelinker clone 1 c= 227,4 ng/µl
- P518= pSB1C3_GFP_RFC25_upper band clone 1 c= 138,3 ng/µl
- P520= pSB1C3_GFP_RFC25_lower band clone 1 c= 141,6 ng/µl
Digestion:
| components | P407 | P516 | P290 | P518 | P520 |
| DNA | 3,5 | 10 | 7 | 10 | 10 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 |
| EcoRI | 0,8 | 0,8 | - | - | - |
| AgeI | 0,8 | 0,8 | 0,8 | - | - |
| NgoMIV | - | - | - | 0,8 | 0,8 |
| SpeI | - | - | 0,8 | 0,8 | 0,8 |
| H2O | 10,9 | 4,4 | 7,4 | 4,4 | 4,4 |
| Total volume | 20 | 20 | 20 | 20 | 20 |
1,0 g Agarose,100 ml TAE (1%), 6 µl GELRED , at 120 Volt
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P407= c= ng/µl
- P516= c= ng/µl
- P290= c= ng/µl
- P518= c= ng/µl
- P520= c= ng/µl
T4 Ligation:
The Ligation was performed as following:
- Vector Volume: µl
- Insert Volume: µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubating for 40 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1blue). The cells were plated on a agar plate with Kana/Cm
Cloning of P5_TATAless downstream of pSB1C3_001_RC_insrepcap_KpnI_back and P5 upstream of pSB1C3_001_RC_insrepcap_KpnI_back
Investigator: Anissa
Digestion of the constructs:
- P320 = pSB1C3_001_RC_insrepcap_KpnI_back c=408 ng/µL
- P320 = pSB1C3_001_RC_insrepcap_KpnI_back c=408 ng/µL
- P180 = pSB1C3_pTAV2 (P5)clone 1 c=108,8 ng/µL
- P184 = pSB1C3_pAAV_RC (P5TATAless)clone 3 c=176,9 ng/µL
| Components | vP320 upstream /µL | vP180 upstream/µL | vP320 downsteam /µL | vP184 downsteam/µL |
| DNA | 2,5 | 13,8 | 2,5 | 8,5 |
| BSA (10x) | 1,5 | 2 | 1,5 | 1,5 |
| Buffer no. 4 (10x) | 1,5 | 2 | 1,5 | 1,5 |
| Enzyme 1 | XbaI 1 | SpeI 1 | SpeI 1 | XbaI 1 |
| Enzyme 2 | EcoRI 1 | EcoRI 1 | PstI 1 | PstI 1 |
| H2O | 7,5 | 0,2 | 7,5 | 1,5 |
| Total volume | 15 | 20 | 15 | 15 |
Loading plan:
M P320(upstream) P320(downstream) P180 P184
Results:
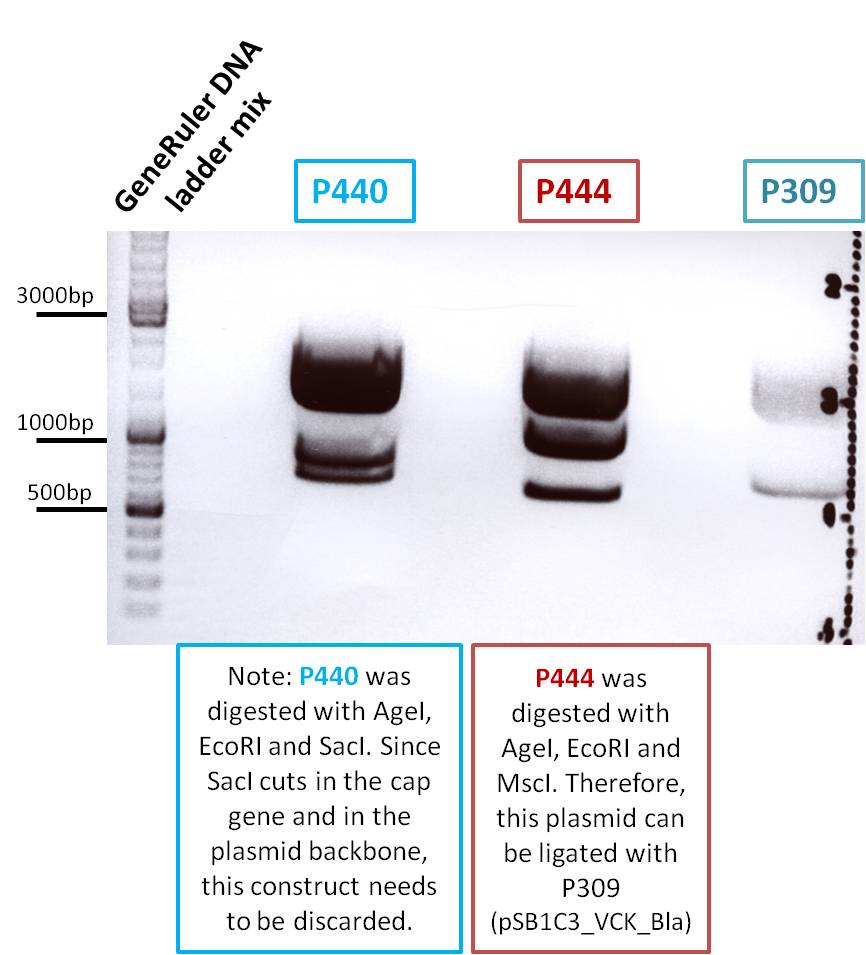
After gel extraction has been performed, the ligation was carried out.
Ligation:
- v=P444 =5,5µL
- v=P309 =2,5µL
The ligation mix was transformed into XL1-Blue cells and plated on agar plates containing chloramphenicol.
Next steps:
Picking clones and perform Mini-Prep. After the Mini-Preps have been performed these constructs can be used for inserting the HSPG knockout motif.
Test digestion of VP2-N-terminal fusion constructs
Investigator: Hanna
Comment: On Satureday a colony PCR was performed of:
1. pCerulean_Zegfr:1907_ShortLinker_VP2/3
2. pCerulean_Zegfr:1907_SEG_VP2/3
3. pCerulean_CFP_MiddleLinker_VP2/3
4. pCerulean_6xHis_MiddleLinker_VP2/3
5. pCerulean_6xHis_MiddleLinker_VP2/3_insCap
6. pCerulean_Zegfr:1907_MiddleLinker_VP2/3_insCap
Because all constructs (4 clones of each approach), including the negative control (pAAV_RFC25), showed positive results, one test digestion of each construct was performed.
Digestion
| components | Components Master Mix /µl |
| BSA (10x) | 7 |
| Buffer 4 (10x) | 7 |
| EcoRI | 3.5 |
| SpeI | 3.5 |
| H2O | 35 |
--> 2 µL DNA + 8 µL Master Mix
- Incubation: 1.5 h
Agarose-Gel:
0.4 g Agarose, 50 mL TAE (0.8 %), 3 µL GELRED, at 100 Volt, running time: 50 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 | Expected size 2 |
|---|---|---|---|---|
| P 530 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 531 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 532 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 533 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 534 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 535 | 10 µl | 2 µl | 2710 bp | 3937 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample ___ /µl | Sample ___ /µl | Sample ___ /µl | Sample ___ /µl | Sample ___ /µl | |
|---|---|---|---|---|---|---|
| Lane |
 "
"